Decode Cellular Senescence and Aging at Single-Cell Resolution
Aging is Among the Largest Predictors of Chronic Disease and Cancer.
Uncovering the underlying mechanisms that drive aging could transform how we prevent and treat many diseases. Yet current approaches face fundamental limitations:
- Too slow: Lifespan endpoints are impractical. Researchers need predictive markers measurable in weeks.
- Too fragmented: No single marker defines aging. Multi-modal hallmarks are more predictive, but rarely measured in the same cells.
- Too narrow: Hallmarks differ across tissues, demanding diverse, high-throughput tools.
What’s needed:
- Multiple hallmarks in one assay: Link morphology, state, secretion, interactions and gene expression in the same cells, over time.
- Structured co-cultures: Reveal how aging or inflamed cells alter healthy populations.
- High-dimensional datasets: Resolve aging into core programs and define druggable mechanisms, opening paths to new therapeutic classes.

Link Hallmarks to Reveal How Aging Processes Unfold.
By integrating morphology, cell state, function and gene expression across tens of thousands of cells, Cellanome enables researchers to resolve senescence, immune dysfunction and inflammation.
The resulting multimodal datasets support selective interventions, open paths to new therapeutic strategies, and can train AI models of tissue decline and repair.
Built to Resolve Aging’s Cellular Complexity

Identify & Track Senescent Cells
Converge multimodal hallmarks in the same single cells; track over days to weeks.
Map Immune Dysfunction
Quantify hallmarks of inflammation, from altered cytokine secretion to impaired microglial phagocytosis.
Track Longitudinally
Monitor specific cells or ensembles over days to weeks; link results to endpoint RNA analyses.

Reveal Interactions
Observe how aging cells influence neighbors, via structured co-cultures.
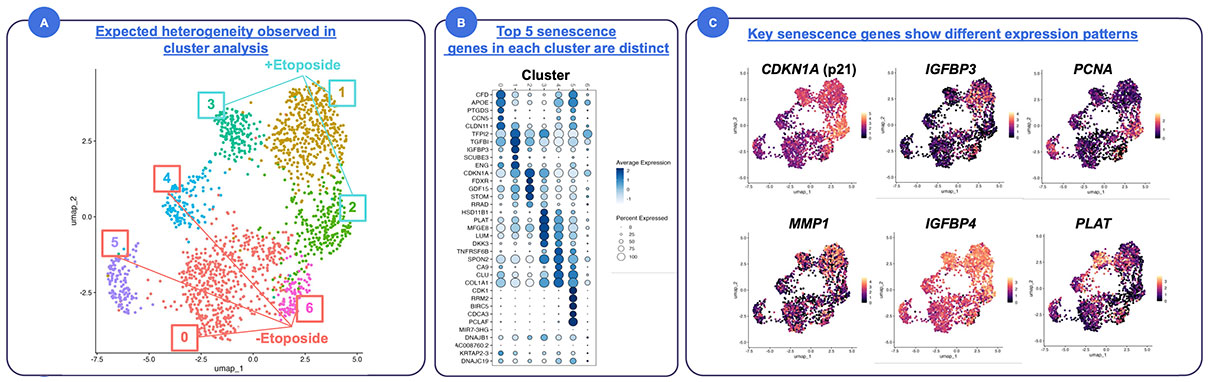
Converging Multi-Modal Hallmarks of Senescence to Single Cells in Preadipocytes
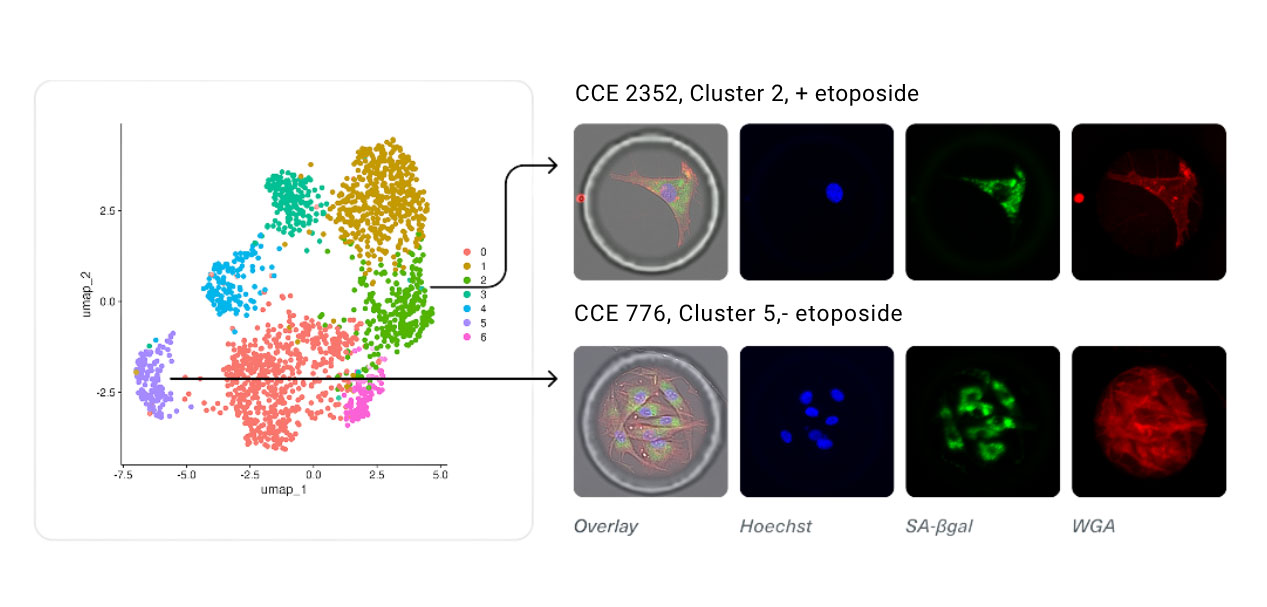
Human primary preadipocytes were cultured in CellCage™ enclosures. Cells were treated with etoposide for 48 hours to induce senescence and compared to untreated control cells. mRNA sequencing showed distinct clusters of cells by gene expression which coincided with proliferative and morphological differences.
Build Disease-Relevant Models. Test Interventions. Accelerate Translation.
Configure disease models with senescence inducers, senolytics, rejuvenation cues, or immune effectors.
Apply perturbations with programmable timing and washout.
Track how specific cells or ensembles respond over time, linked to RNA and CRISPR perturbations for mechanistic insights.
Generate multimodal, time-resolved datasets to support lead optimization, patient stratification, and AI training.
Build Scalable Models for Disease Research
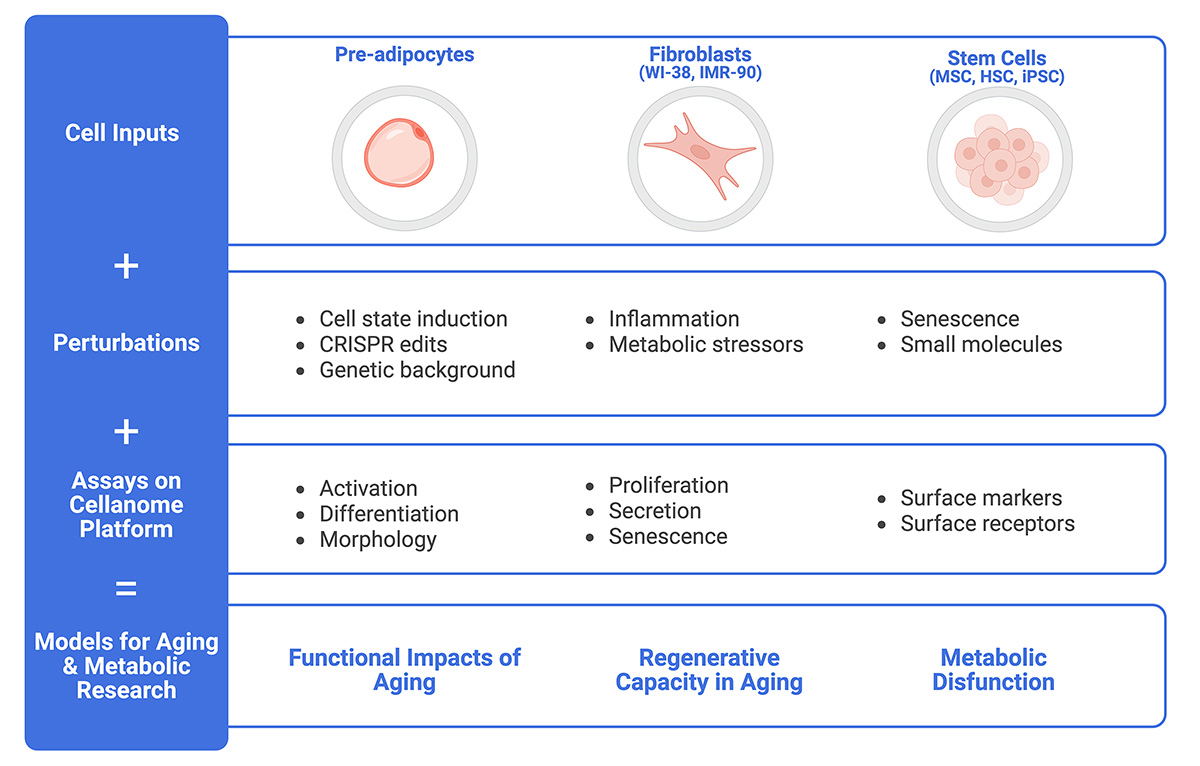
The Cellanome platform supports a variety of cell types and numerous assays, making it easy to create longitudinal models of aging and metabolic disfunction.
From Mechanisms to Therapeutics

Resolve states. Quantify signals. Test interventions.
- Identify senescent cells with confidence: Distinguish them from quiescence or transient stress responses by linking multiple hallmarks in the same cells. Resolve gradual state changes.
- Resolve immune dysfunction: Capture hallmarks of inflammation such as cytokine signaling, microglial clearance defects, and glial remodeling.
- Interrogate therapeutic response: Reveal on‑target mechanisms, off‑target effects, and early toxicity.
- Generate training data. Feed AI with structured, time-resolved maps of state, secretion, and drug response–all precisely resolved to individual cells or interacting ensembles.
Exploring the Frontiers of Aging Biology
Leading labs are moving beyond population-level markers to single-cell, time-resolved approaches.
Case Study: Multi-Modal Identification of Senescent Cells in Human Preadipocytes
Senescent cells can now be defined with higher confidence by converging multiple hallmarks within the same cells.
Senescence is central to aging and age-related diseases, but progress has been limited because no single marker is specific. Conventional assays split samples across modalities, yielding “senescent-like” populations that are likely impure, making it challenging to employ fundamental cell biology approaches (e.g., cell quantification, co-cultures).
Approach:
Using the Cellanome R3200 platform, researchers:
- Enclosed thousands of human preadipocytes in CellCage enclosures.
- Induced senescence with etoposide.
- Tracked each cell’s morphology and proliferation over 5 days.
- Linked phenotypes to SA-β-gal, p21, cytokine secretion , and single-cell transcriptomics.
What they found:
Distinct subpopulations with different combinations of hallmarks. Transcriptomics confirmed senescence pathways and revealed shared and divergent gene programs across clusters.
Why this matters:
This approach provides a framework for increasing confidence in identifying senescent cells. By reducing ambiguity, it opens the door to:
- Discovery of tissue-specific markers within aging atlases.
- More selective senolytics that spare healthy cells.
- Co-culture studies on how primary and secondary senescence propagates.
- Potential biomarkers to track senescent burden and clearance.
- High-throughput drug screening against defined senescent subtypes, accelerating discovery of safer, more effective therapies.