
Reveal How Interactions Shape Immune Responses, Pathology, and Therapeutic Efficacy
Immune Protection and Pathology Arise From Cell Interactions
- Immunity is driven by cell-cell interactions. Diverse cells sense their environment, signal to one another, and act in coordination. Protection, pathology, and therapeutic response all emerge from these exchanges.
- Interactions are heterogeneous. The same cell type can prime or suppress, sustain activation or enter exhaustion. Even within the same task, such as killing or activation, cells vary in how effectively or persistently they perform. These layers of variability influence whether immunity protects or drives pathology.
- No single measure captures this complexity. A cell that secretes cytokines may not also be a sustained killer. Cells with similar RNA profiles can diverge in function depending on context.
- Most workflows capture fragments. Static snapshots, single readouts, or population averages. It remains very challenging to follow defined ensembles over time, at scale, across multiple layers of function.

Resolve How Immune Ensembles Signal, Act, and Adapt.
With Cellanome, immunologists can model defined ensembles, perturb them, and track which specific interactions persist or fail. By linking multiple live-cell behaviors to RNA programs across tens of thousands of interacting ensembles, the platform can resolve the ambiguities of fragmented measurements.
Capabilities of the Cellanome R3200 in Immunology
- Model cell-cell interactions: Co-culture T-cells, B-cells, dendritic cells, macrophages, and relevant targets across tens of thousands of ensembles in parallel.
- Preserve cell adherence: Maintain DCs, macrophages, fibroblasts, and adherent tumor cell targets in their natural attached state, preserving morphology, function and viability.
- Apply perturbations: Introduce cytokines, antibodies, and small molecules with programmable timing and washout.
- Track longitudinal dynamics: Monitor proliferation, killing, secretion and state transitions (activation, exhaustion, tolerance) over time.
- Link to RNA programs: Link differential outcomes to RNA signatures and CRISPR edits within the same interacting cells, resolving mechanisms of immune activation, suppression, and dysfunction.
Configure Immune Ensembles. Test Perturbations. Reveal Mechanisms.
One system, many models. Build flexible disease-relevant immune ensembles and test perturbations in contexts that currently require multiple assays, accelerating the path from mechanistic discoveries to therapeutic relevance.
Build Scalable Models for Disease Research
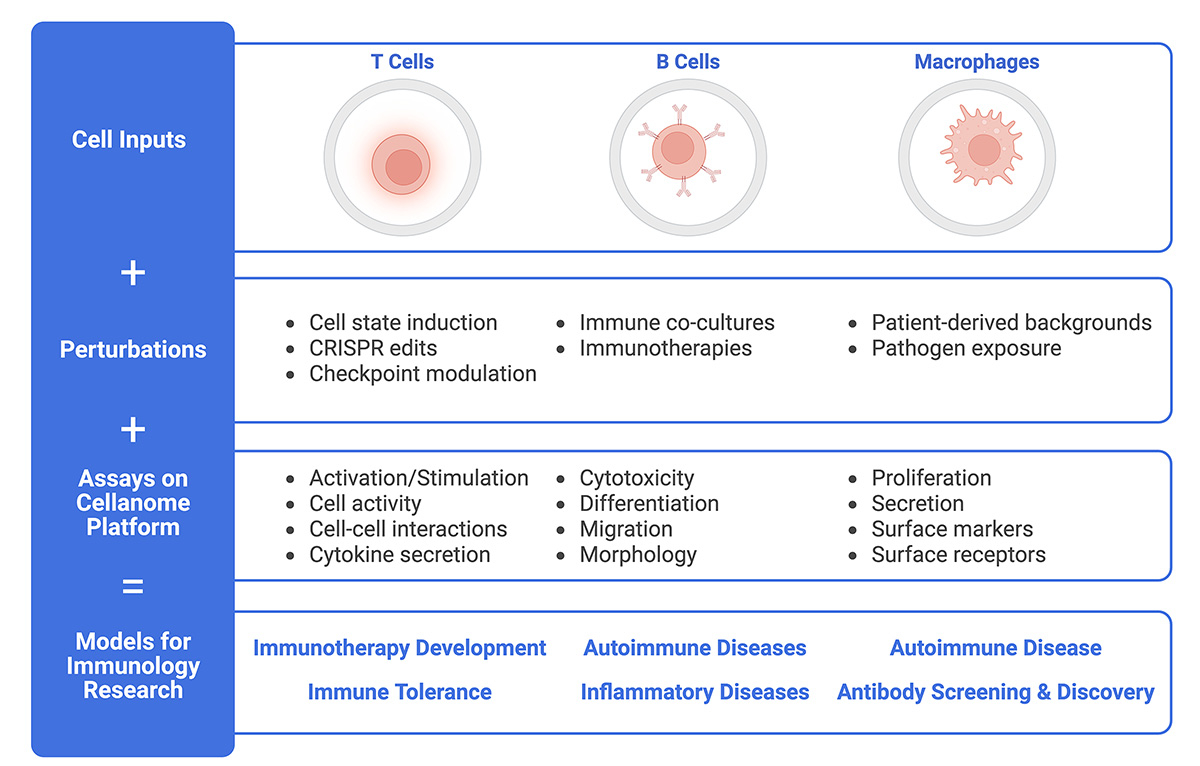
The Cellanome platform supports a variety of cell types and numerous assays, making it easy to create longitudinal models for investigating immune-related diseases.
From Cellular Mechanisms to Durable Therapies

The Cellanome Platform links structure, function, and gene expression across time - at a fidelity and scale unmatched by any other platform. It opens a new tier of mechanistic insight, ready for screening, modeling, and drug development.
- Resolve heterogenous interactions: Track how specific engagements diverge in outcome - priming, tolerance, or exhaustion.
- Interrogate transient vs durable responses: Distinguish short-lived events from programs that sustain protection or drive pathology.
- Connect function to mechanism: Tie differential outcomes to RNA programs to uncover pathways that underlie immune response, resistance, or dysfunction – and new therapeutic targets.
- Guide therapeutic strategy: Couple these insights with perturbations to help design combinations, uncover biomarkers of durable immunity, and guide the design of safer, more effective interventions.
Exploring the Frontiers of Immunology
New capabilities unlock new questions. These case studies show how scientists are advancing immunology research with Cellanome.
Case Study: Extending CRISPR Screens Beyond Single Cells to Ensemble-Level Biology: Linking DC Perturbation to T-Cell Activation
CRISPR-edited dendritic cells were co-cultured with OT-I T cells to map how gene knockouts alter priming capacity under tumor-driven immune-suppression.
Researchers used the Cellanome R3200 to develop an assay that explores modulators of T cell priming in dendritic cells (DCs) impaired by cancer-induced immunosuppression. CRISPR-edited DCs were co-cultured with one or more OT-I T cells to map how gene knockouts altered priming. Imaging readouts of T-cell proliferation and activation were tied to sgRNA detection and mRNA.
This study delivers two advances:
First, by linking DC edits to T-cell activation under immune-suppressive conditions, it provides a model for studying how tumors disable antigen presentation, and how this might be reversed.
Second, it demonstrates a framework that expands high-throughput CRISPR screens beyond single-cell states (e.g., what happens inside a single cell) to ensemble-level phenotypes (e.g., how perturbations in one cell reshape the function of interacting cells).
In this case, DC edits were linked to downstream T-cell activation, but the principle applies broadly to tumor-immune interactions, neural pruning, or any system where outcomes are defined by cell-cell interactions.
What’s next:
Develop an immunosuppressive culture model that mirrors the cytokine milieu DC’s experience in vivo during tumor progression, and test which knock-outs rescue priming.
Extend beyond immune-oncology to fibrosis or neuro-immune interactions, where functions and outcomes are also defined by ensembles.
Learn More About Cellanome
Ready to Connect Cell Behavior to the Programs that Drive It?
Contact us to learn how the Cellanome Platform can transform your immunology research.

