Uncover Neural States, Networks, and Dynamics at Scale
Neural Biology is Demanding
Neurobiology research places high requirements on analytical tools.
- Fragile, adherent cells. Neurons, astrocytes, and microglia lose structure in suspension. Enzymatic dissociation severs contacts and distorts morphology.
- Dynamic processes. Differentiation, neurite outgrowth, and glial response unfold over hours to weeks.
- Coordinated function. These cells don’t act alone. They signal, adapt, and repair in networks.
- Conventional workflows break the chain. Functional outputs are rarely linked to transcriptomic information with conventional assays. Many labs lose over 50% of cells during harsh transfer steps across multi-step workflows.

Understand How the Brain Responds, Repairs, and Rewires.
Track live interactions across structured neural ensembles, reveal transcriptomic programs that drive resilience or failure, and connect perturbations to functional outcomes with mechanistic insights.
In one continuous experiment, you can culture neural cells, track their interactions over time, and uncover the transcriptomic programs that shape repair, degeneration, and response.
- Adherent Cell Support: Culture and assay CNS cells directly on the flow cell, preserving morphology, viability and function.
- Defined Ensembles: Study individual cells or multi-cellular units (e.g. neuron–glia pairs, neurospheres) inside permeable CellCage™ enclosures, with the option to build co-cultures over intact neuronal networks.
- Time-Resolved Interactions at Scale: Track dynamic morphotypes and live-cell processes (eg, phagocytosis, neurite extension) over days to weeks, across tens of thousands of CellCage enclosures (CCE's).
- Link Perturbation to Function to Transcriptome: Gate by function or morphotype, and resolve RNA programs that drive differential outcomes.
- Integrated Workflow, Fewer Transfer Steps: Preserve sample integrity and minimize technical noise by performing multiple assays (fluorescence-based read-outs and RNA) within a unified experimental framework.
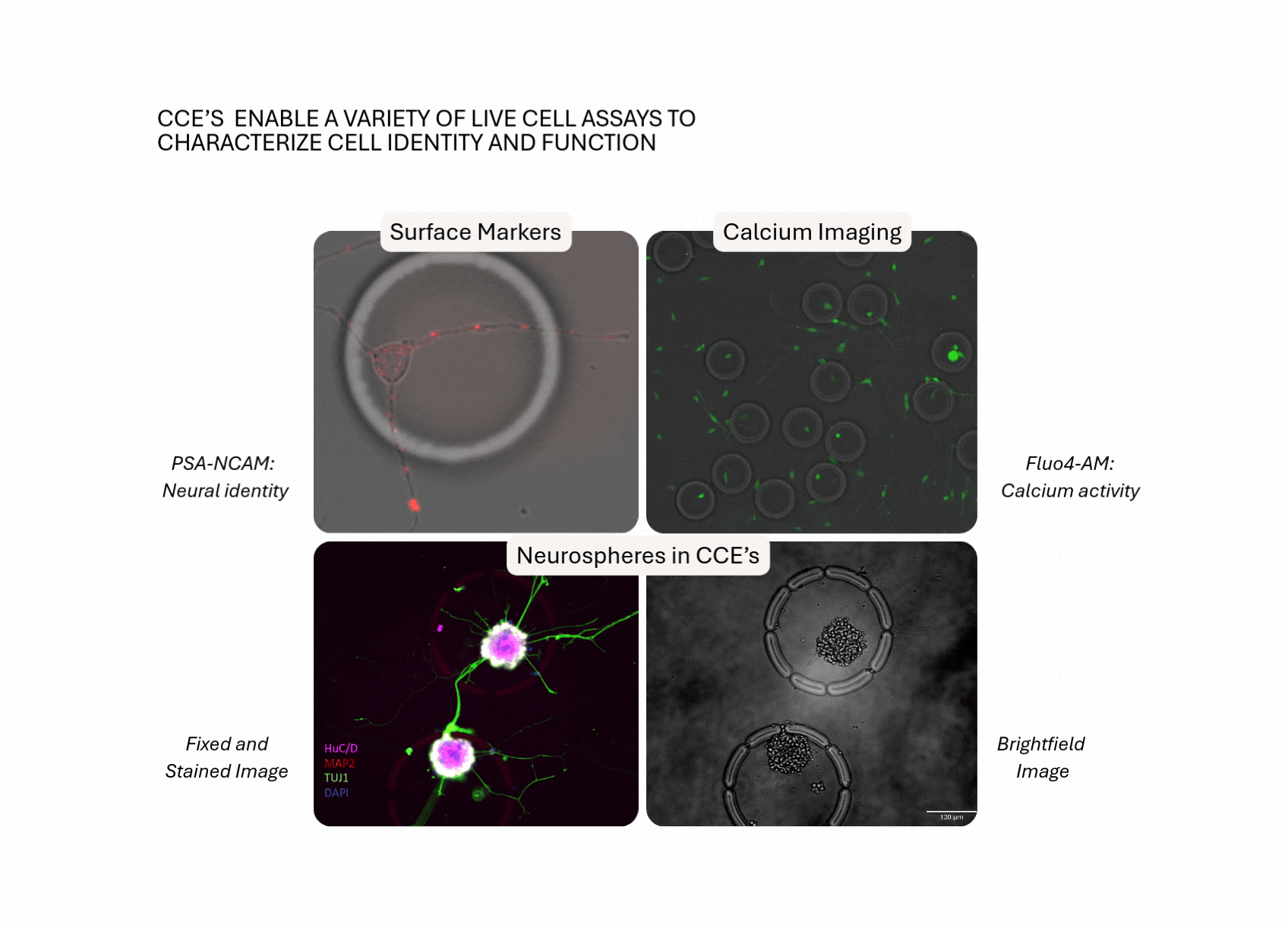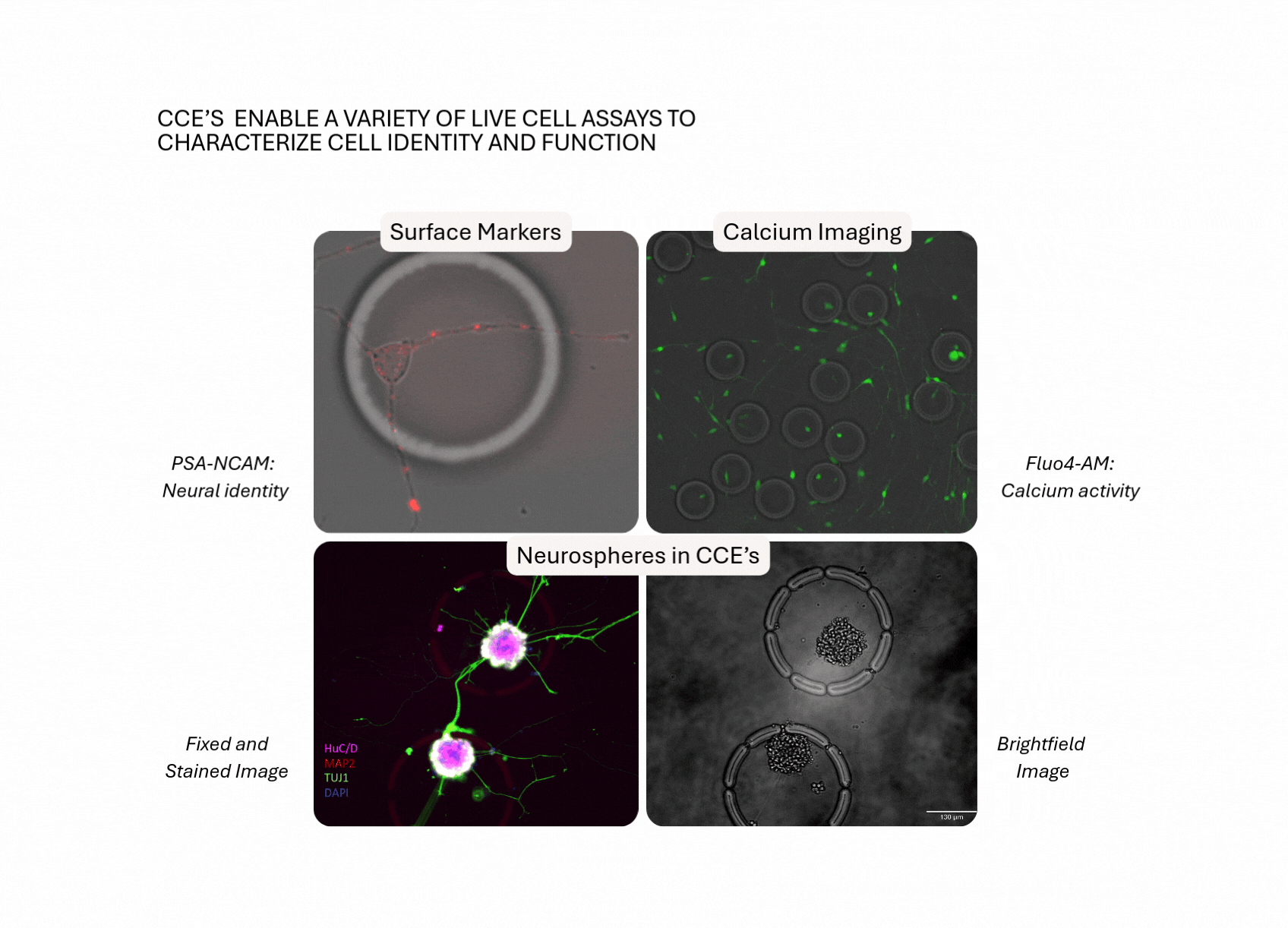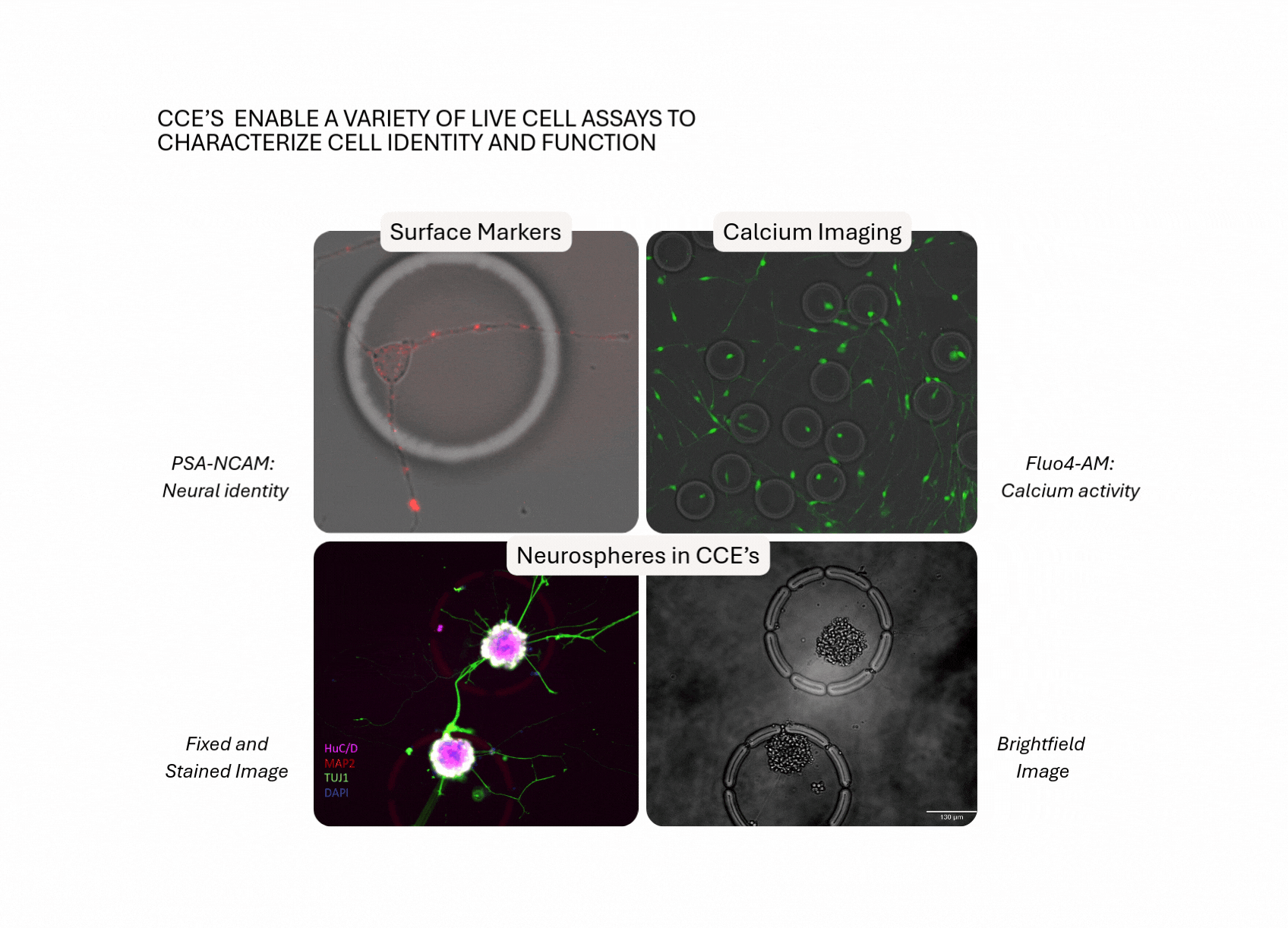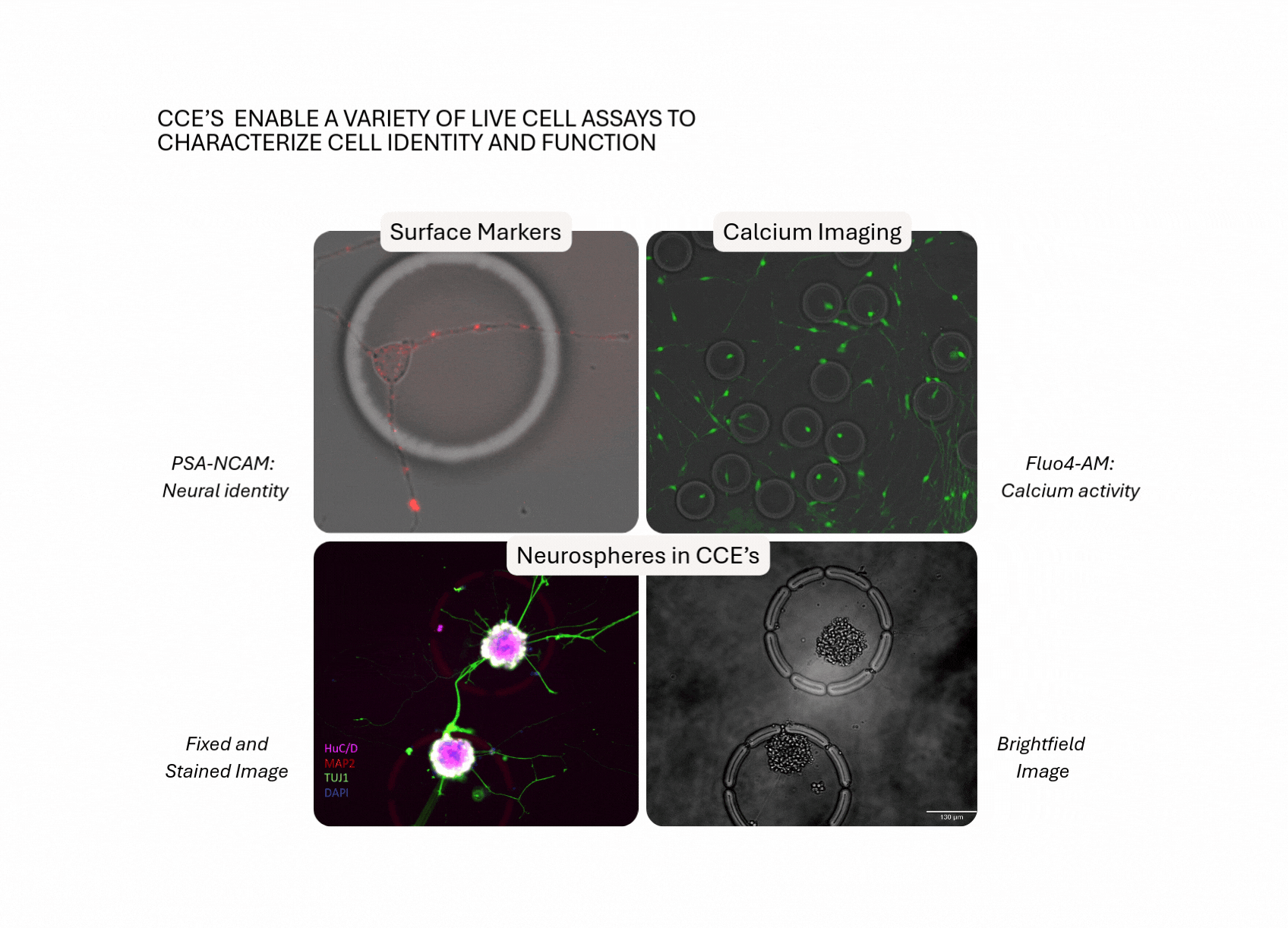
Build Disease-Relevant Models. Test Perturbations. Reveal Mechanisms.
Treat CNS-relevant cells with small molecules, cytokines or antibodies at programmed times to simulate dosing regimens.
Track how ensembles respond over time, and link those behaviors to transcriptome programs and CRISPR guide expression.
Generate multimodal, longitudinal datasets to support lead optimization, combination strategies, and patient stratification or to train deep-learning models on true biological complexity.
Build Scalable Models for Disease Research
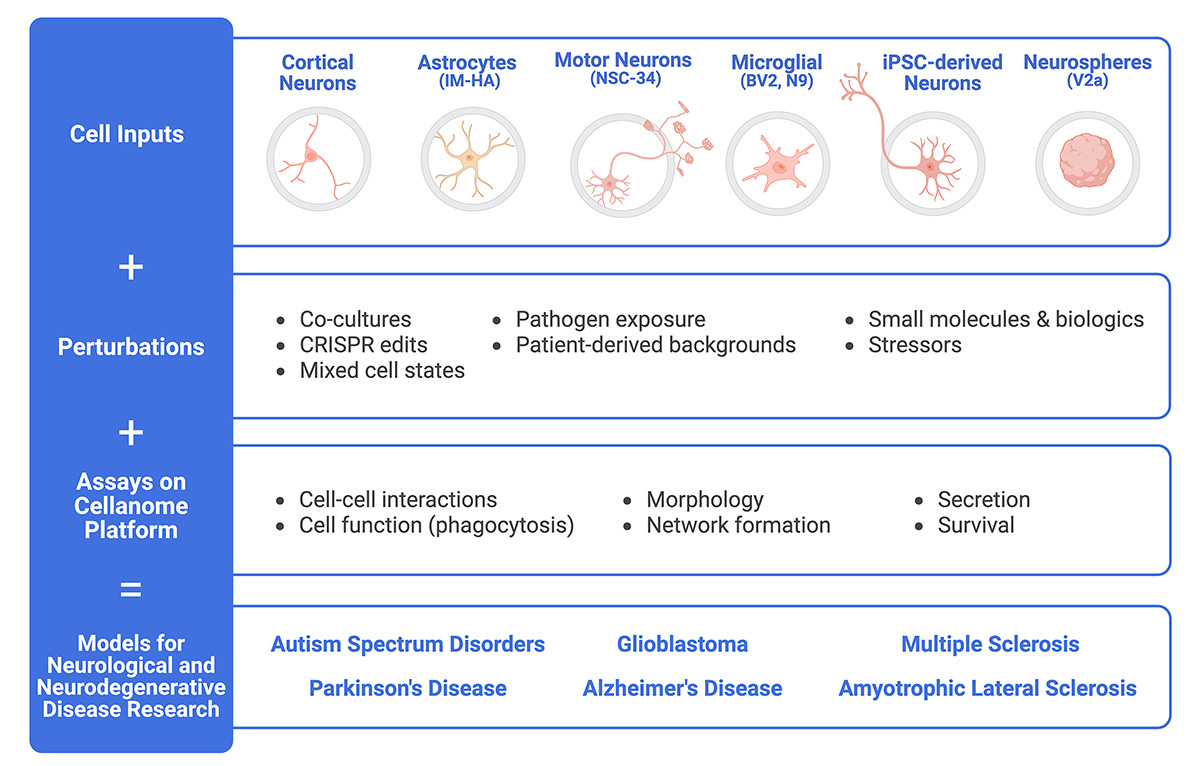
The Cellanome platform supports a variety of cell types and numerous assays, making it easy to create longitudinal models for studying neurological and neurodegenerative diseases.
Unveiling Dynamic Cellular Choreography in Neurobiology

New experiments. New signals. New possibilities.
The Cellanome platform links structure, function, and gene expression across time and at a fidelity and scale unmatched by any other platform. It opens a new tier of mechanistic insight, ready for screening, modeling, and drug development.
- Track how resilience emerges. Follow glial response to stress or stem cells differentiating into neurons across morphology, function, and gene expression.
- Model complex diseases. Observe transitions and interactions over time. Study the behaviors of different cell combinations and uncover context-specific genetic programs behind these differences.
- Dissect therapeutic responses. Test how small molecules, antibodies, or genetic perturbations/edits shift cell states and network behavior. Reveal mechanisms, off-target effects, early signs of toxicity, or opportunities for combination therapies.
- Train models with biological fidelity. Feed AI systems with data that reflects true CNS complexity; dynamic, multimodal, ensemble-based, and time-resolved.
Exploring the Frontiers of Neurobiology
New capabilities unlock new questions. These case studies show how scientists are exploring neural complexity with Cellanome.
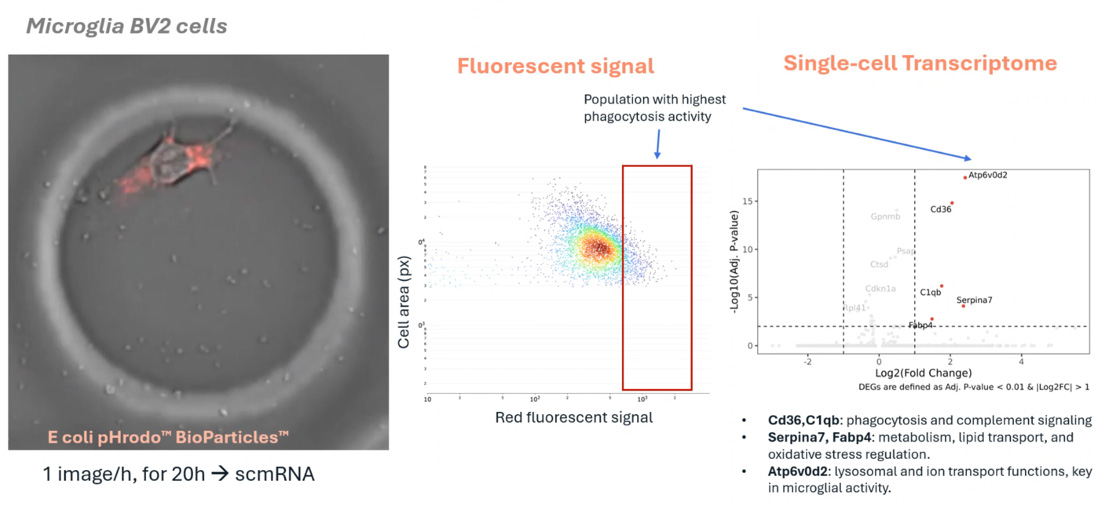
Case Study: Functional Profiling of Microglia in Neuroinflammation
Link microglial behavior to gene expression at single-cell resolution, for insight into neuroinflammation, drug response, and immune dysfunction in CNS disease.
Researchers used the Cellanome R3200 to enclose individual microglia with fluorescent particles and track phagocytosis over 12 hours via fluorescent imaging. Each cell’s transcriptome was then sequenced, linking activity levels to gene expression.
What they found:
High-activity microglia upregulated genes in complement signaling, lipid metabolism, and lysosomal function–key pathways in neuroinflammation and repair.
Why it matters:
This approach overcomes key limitations in standard assays by capturing phagocytic function and gene expression in the same individual cells without dissociation, pooling, or inference. It enables a direct, scalable readout of immune heterogeneity, and reveals the transcriptional programs driving effective or impaired microglial responses.
What’s next:
Extend to co-cultures by layering enclosed microglia over intact neuronal networks. Study how cell-cell interactions shape phagocytic behavior and fate. Combine with cytokines, CRISPR libraries, or immunotherapies to generate time-resolved, multi-modal datasets that can be used for MoA analysis, early biomarker discovery, and AI-guided modeling in CNS disease.
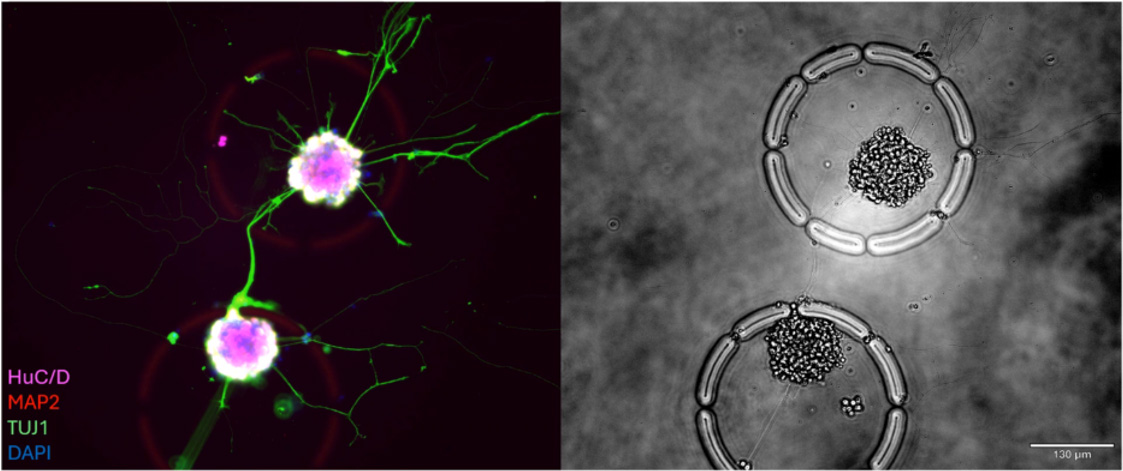
Case Study: Modeling Synapse Formation and Developmental Trajectories in 3D
Track development, function, and gene expression in intact neurospheres, a human-relevant 3D model increasingly vital as regulators move away from animal studies.
Stem-cell-derived neurospheres offer a robust 3D model of early brain development, but standard assays disrupt their structure and miss critical dynamics.
Approach:
Using the Cellanome R3200, the research team explored,
- Hundreds of intact neurospheres (100–200 cells each) were cultured inside individual CellCage™ enclosures.
- Axon extension, synapse formation and calcium activity were tracked over multiple days.
- End-point RNA-Seq was linked back to each neurosphere’s functional behavior.
- UMAP clustering revealed lineage-specific gene programs, validated by fluorescent markers.
What's next:
This lays the groundwork for CRISPR-based multimodal screens to probe mechanisms of development, degeneration, and repair within preserved 3D architecture.
Why it matters:
As the FDA and others move to reduce reliance on animal models, human-relevant in vitro systems like this are increasingly essential.
Learn More About Cellanome
Ready to Uncover Neural States, Networks, and Dynamics at Scale?
Contact us to learn how the Cellanome Platform can transform your neurobiology research, from target discovery to patient impact.