Link Tumor–Immune Interactions to RNA Programs, at Scale
Cancer is Defined by Heterogeneity and Shaped by Interactions.
- Tumor cells exhibit profound molecular diversity. Beyond genetic mutations, tumor cells enter cryptic states shaped by post-transcriptional regulation, protein activity, and signals from nearby cells.
- Morphology and transcriptome can mislead. Cells that look alike may diverge in molecular and functional state, while transcriptomically similar cells may behave differently.
- Interactions amplify complexity. Immune and stromal cells compound this diversity, driving emergent and multifaceted resistance mechanisms.
- These dynamics drive therapeutic outcomes. They help to explain why patients respond differently, why treatments lose durability, and why serious adverse events remain common.
- Tools must keep pace. Progress requires platforms that can model defined cell communities, perturb their interactions, and track evolving responses over time. These include: resistant clones, persister states, and immune shifts that single-timepoint or single-modality assays miss.
Map the Cellular Choreography That Drives Response and Resistance.
We’re building tools that reveal tumor–immune interactions as they unfold, making it possible to link differential responses to the RNA programs that drive them.
Capabilities of the Cellanome R3200 in Cancer Research
- Model tumor communities: Build co-cultures of tumor cells with T-cells, macrophages, or fibroblasts. Preserve adherent growth for solid tumor models, avoiding the need to study cells in suspension, which can alter function and viability.
- Perturb co-cultures: Apply targeted agents or immunotherapies with precise timing and washout to probe cytotoxicity, persistence, and relapse.
- Track longitudinal dynamics: Monitor proliferation, killing, recurrence, and clonal outgrowth over days to weeks, capturing transitions that single snapshots miss.
- Resolve mechanisms: Link differential outcomes to RNA signatures and CRISPR guide expression, revealing pathways of resistance, immune dysfunction, and tumor-stroma cross-talk that can guide biomarker discovery and therapeutic strategy.
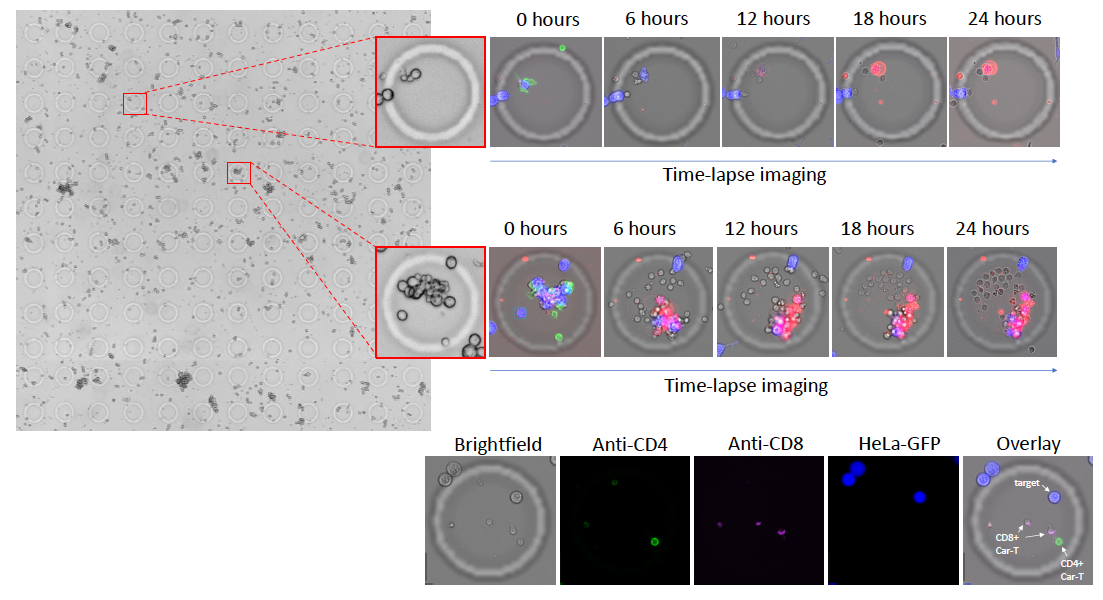
Identification of T-Cell Types and Longitudinal Monitoring of Cell Killing Activity Using AIC100 CAR T-Cells

AIC100 cells, labeled with anti-CD4 (green) and anti-CD8 (violet) antibodies were co-encapsulated in CellCage™ enclosures with HeLa-GFP target cells (blue). Cell killing, as indicated by Annexin V staining (red), was monitored for each CellCage™ enclosure via time-lapse imaging over 24 hours.
Build Disease-Relevant Models. Test Perturbations. Reveal Mechanisms.
Cellanome’s flexible assays enable researchers to configure disease-relevant co-cultures, apply perturbations with precise timing and washout, and capture outcomes ranging from proliferation and cytotoxicity to persistence and relapse across tens of thousands of parallel enclosures.
RNA programs can be resolved at the level of specific cells or ensembles and linked to the preceeding longitudinal measurements, enabling mechanistic insights that support biomarker discovery, therapeutic evaluation, and rational combination design.
Build Scalable Models for Disease Research
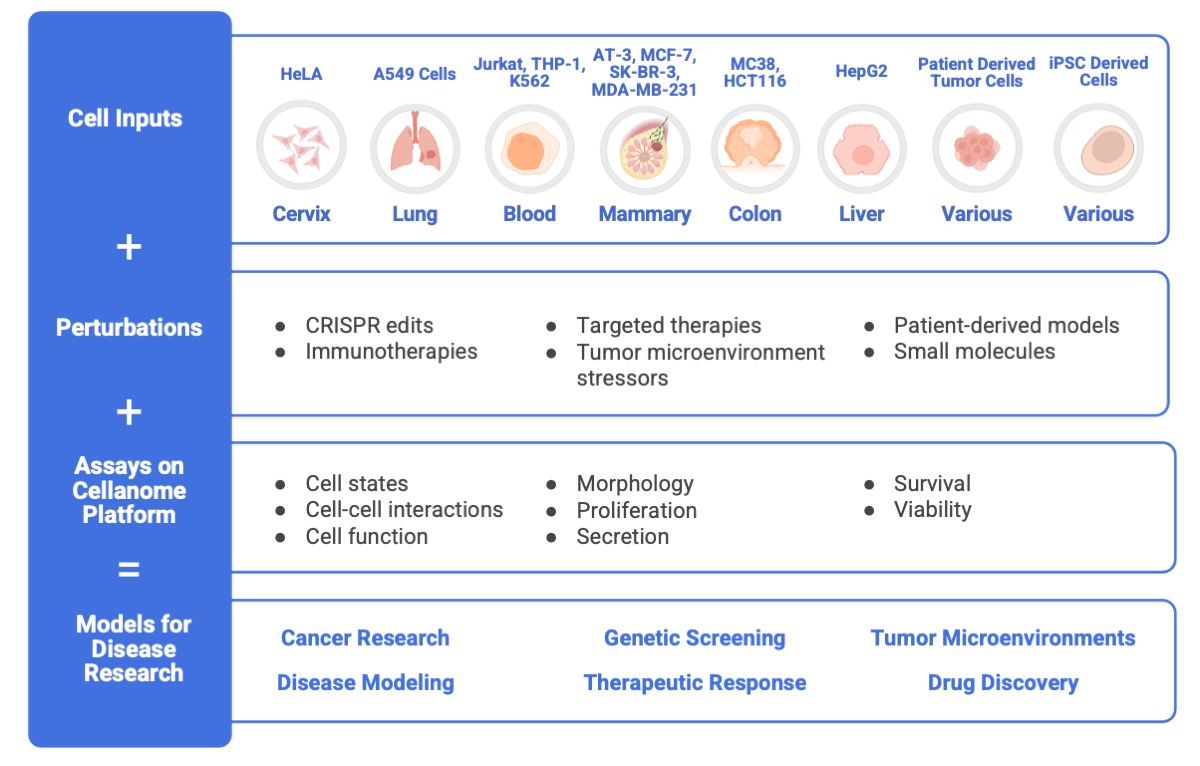
The Cellanome platform supports a variety of cell types and numerous assays, making it easy to create longitudinal models for investigating disease mechanisms and powering drug discovery.
From Cellular Mechanisms to Durable Therapies.
Dynamic Co-Cultures. Mechanistic Clarity. Durable Therapies.
Response in cancer is defined by how cells adapt together. Cellanome resolves these interactions at scale and ties outcomes to RNA programs, enabling the mechanistic clarity needed to design more durable treatments.

- Resolve resistance programs: Distinguish true resistance from transient stress responses, identify persister states, and track clonal outgrowth over time
- Track tumor–immune dynamics at scale: Measure cytotoxicity, immune evasion, and stromal signaling across tens of thousands of defined co-cultures.
- Resolve MoAs and evaluate therapeutic strategies: Track how edits or treatments affect function, morphology, and gene expression in adherent cells or interacting ensembles. Reveal on-target mechanisms, off-target effects, and context-dependent responses.
- Enable predictive models: Generate longitudinal, multimodal datasets that inform biomarker discovery, guide therapeutic strategy, and train AI models to predict response and relapse.
Exploring the Frontiers of Immune-Oncology
Leading scientists are exploring new methods of interrogating immune cell biology with Cellanome.
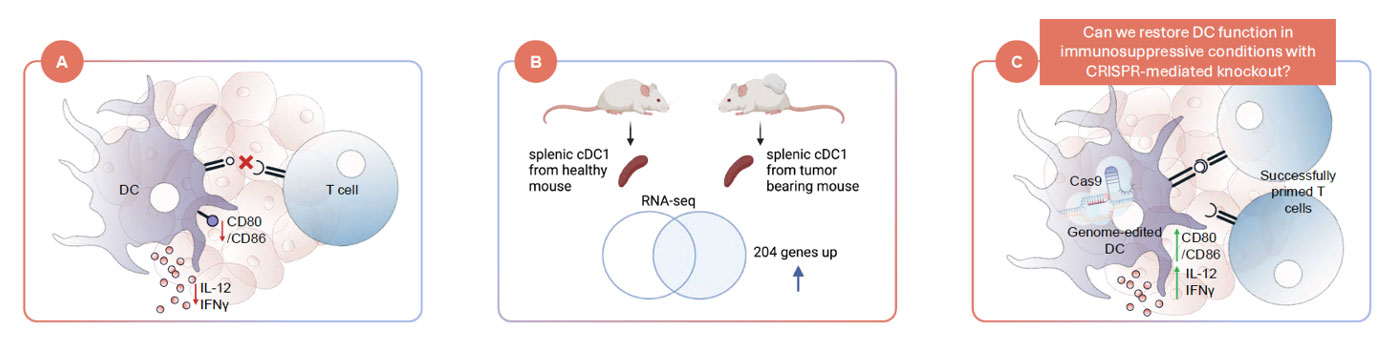
Case Study: Enabling Cell-Extrinsic Read-Outs for CRISPR Screening - Identifying Dendritic Cell Pathways that Regulate T Cell Priming
CRISPR-edited dendritic cells were co-cultured with T cells to identify genes that may restore T-cell activation under tumor-driven immune-suppression conditions.
Approach:
Researchers used Cellanome’s R3200 to develop an assay that explores modulators of T cell priming by dendritic cells (DCs) impaired by tumor-induced immunosuppression.
CRISPR-edited DCs were co-cultured with one or more OT-I T cells to identify gene knock-outs that may increase T-cell priming. Alterations of priming activities, as monitored by T cell proliferation and activation, were linked to sgRNA identification and transcriptomics.
What they found:
Linking DC CRISPR edits to T-cell priming under immune-suppressive conditions, it provides a model for studying how tumors disable antigen presentation, and how this might be reversed.
The study demonstrates a framework that expands high-throughput CRISPR screens beyond cell-intrinsic read-outs (i.e., what happens inside a single cell) to cell-extrinsic read-outs (i.e., how perturbations in one cell reshape the function of interacting cells).
In this case, DC edits were linked to downstream T-cell activation, but the principle applies broadly, including to tumor-immune cell interactions, neural pruning, or any system where outcomes are defined by cell-cell interactions.
What’s next:
Develop an immunosuppressive culture model that mirrors the cytokine milieu DCs experience in vivo in the tumor microenvironment (TME), and test which knock-outs rescue T-cell priming.
Extend beyond immune-oncology to fibrosis or neuroimmune interactions, where functions and outcomes are also defined by ensembles.
Case Study: Multi-Modal Characterization of CAR-T Cytotoxicity
Multi-modal profiling reveals how CAR-T subsets interact to drive potency and durability.
Cell therapies have transformed the treatment of hematologic cancers, but solid tumors remain challenging. In vivo efficacy is hard to predict, product heterogeneity complicates interpretation, and current assays often require stitching together disparate datasets.
Approach:
Using the Cellanome R3200 Platform, researchers investigated:
Tens of thousands of CAR-T (AIC100, Phase 1 thyroid cancer) and TALL-104 cells were co-cultured with targets in CellCage enclosures.
Time-lapse imaging integrated cytotoxicity, proliferation and cytokine secretion alongside surface markers profiling.
What They Found
The platform resolved cytotoxic subsets invisible to bulk measures: strong, moderate, and weak killers. For AIC100, CD4⁺ and CD8⁺ CAR T-cells showed distinct killing behaviors, and mixtures outperformed isolated subsets, suggesting cooperative mechanisms that drive potency.
Why it Matters
This establishes a framework for defining potency mechanistically, identifying which cells, in which combinations, drive durable therapy, and for guiding next-generation CAR-T design.