
Resources: Presentations
Decoding Immune Cell Interactions: Dr. Matt Spitzer's Approach to Understanding Tumor-Induced Immune Dysfunction
Presented by:
Matthew H. Spitzer, Ph.D.
Immune responses don’t emerge from individual cells acting alone. They emerge from interactions, unfolding over time and shaped by context. Whether immunity protects, fails, or becomes pathological depends on how these interactions play out.
Dr. Matt Spitzer’s lab at UCSF studies these interactions to understand why immune-based cancer therapies succeed in some patients but fail in others. In this talk, he describes experimental approaches his lab is using to directly connect immune cell interactions to functional outcome and RNA programs.
The Biological Problem: When Tumors Disrupt Immune Priming
A central focus of Spitzer’s lab is T-cell activation, which depends on a sequence of interactions:
- Dendritic cells (DCs) recognize the pathogen
- DCs secrete cytokines and physically engage T cells to prime them
- Activated T cells must then find and kill tumor cells
In tumor-bearing mice, Spitzer’s team observed a striking failure of this process. When challenged with a bacterial vaccine, these animals mounted markedly impaired T-cell responses, driven by dysfunctional dendritic cell priming. This has direct clinical implications, as many cancer immunotherapies rely on vaccines to generate anti-tumor T-cell responses, yet patients often fail to respond.
“If the patient is refractory to responding to that vaccine in the first place because their immune system has been dysregulated by the presence of the cancer, of course this therapy won’t work. And this is what we unfortunately have experienced quite commonly in the clinic.”
The core question became: "How do tumor cells suppress dendritic function, and how can that dysfunction be reversed?”
Why Traditional Methods Fall Short
Traditionally, processes governed by cell interactions have been difficult to study.
“We have great tools to study individual cells. […] And then we have really low throughput ways to study the outcome of interactions between these cells. We could, say, take a tumor section and measure how many dying tumor cells are there. Or we could measure tumor shrinkage by actually physically measuring the size of a tumor in a mouse or a human. But these are very low throughput, and they really limit our ability to link the molecular response to this ultimate physiological response.”
Spitzer’s team initially pursued a strong mechanistic hypothesis around a specific receptor based on single cell data, spent considerable time building a knockout mouse, and tested it carefully. The result was disappointing.
“There was absolutely no difference in the ability of those dendritic cells to respond.”
“That’s the classic way we would have approached the problem in the past, but we needed an alternative”, Spitzer explains, “and so this is where we really turned to the Cellanome platform”.
RNA sequencing had previously revealed ~200 genes dysregulated in dendritic cells from tumor-bearing mice, with similar patterns in human samples. They designed a genetic screen with the hopes of identifying causal genes that change the DCs ability to engage with the T cell.
Spitzer acknowledges existing single cell methods could measure cell intrinsic proxies, such as surface markers like CD80. “But the challenge is that CD80 is only loosely related to the ultimately phenotype that we care about, which is T-cell priming.”
“Ideally, what we would do is edit the gene in the dendritic cells and then read out whether it restores the T cell response. And that’s been a challenge historically.”
A New Experimental Paradigm: Cell Extrinsic CRISPR Screens
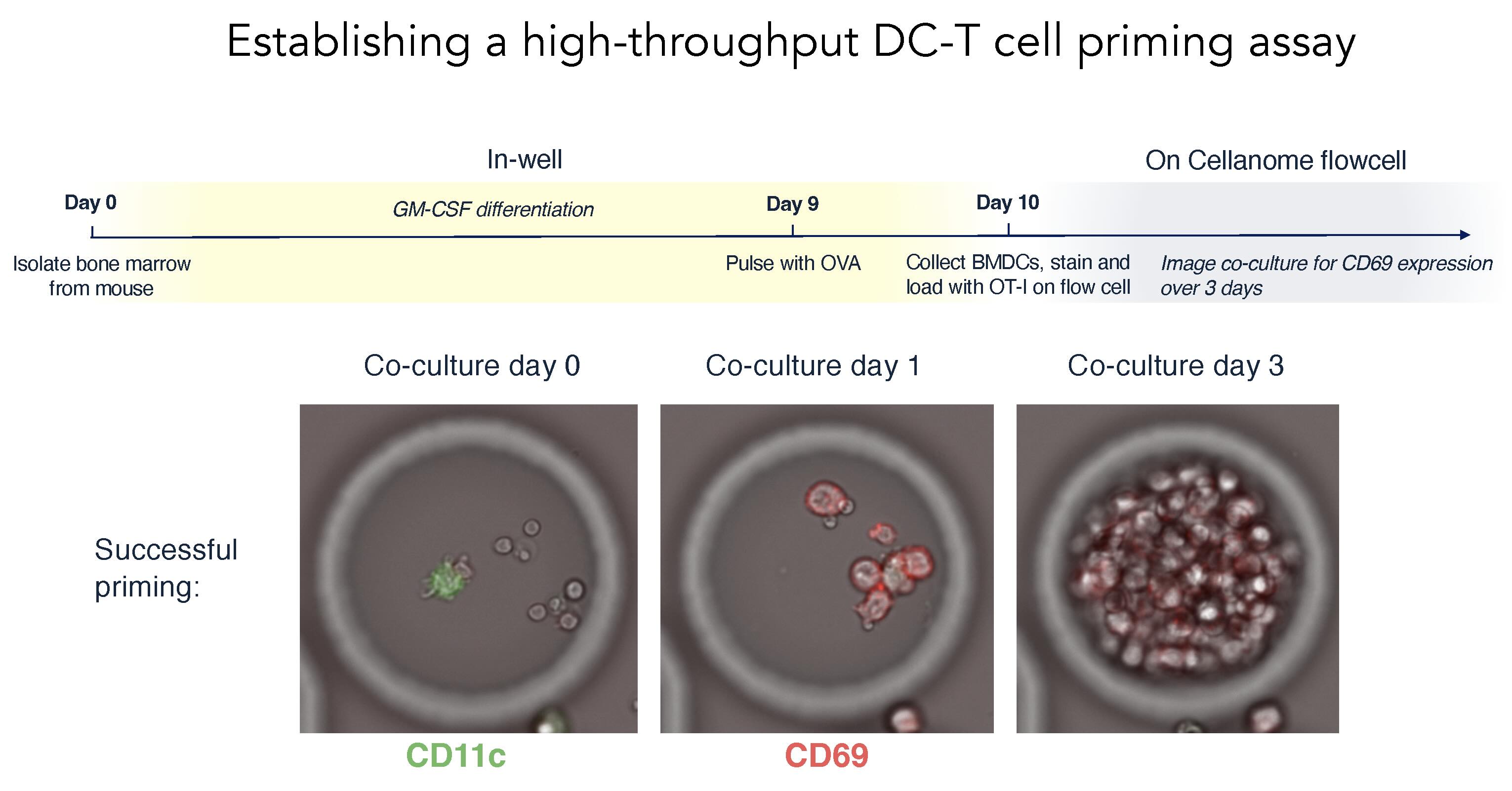
To address this gap, Spitzer's lab developed a new approach using Cellanome’s R3200 platform:
- CRISPR-edited dendritic cells are paired with one or more T cells inside individual CellCage enclosures.
- Over three days, live-cell imaging tracks T cell activation and proliferation.
- At the end, RNA-Seq and guide RNA identity are captured from each dendritic cell.
This enables a direct link between genetic perturbation, dendritic cell behavior, and T-cell response, including under tumor-derived suppressive conditions.
“That’s where we’ve gotten really excited about opportunities that Cellanome creates to really examine in detail the molecular choreography that takes place between these cells during this fundamental decision-making process.”
The approach revealed striking heterogeneity. Even among seemingly identical dendritic cells from healthy individuals, only a subset showed the protein expression profiles associated with strong T cell priming capacity.
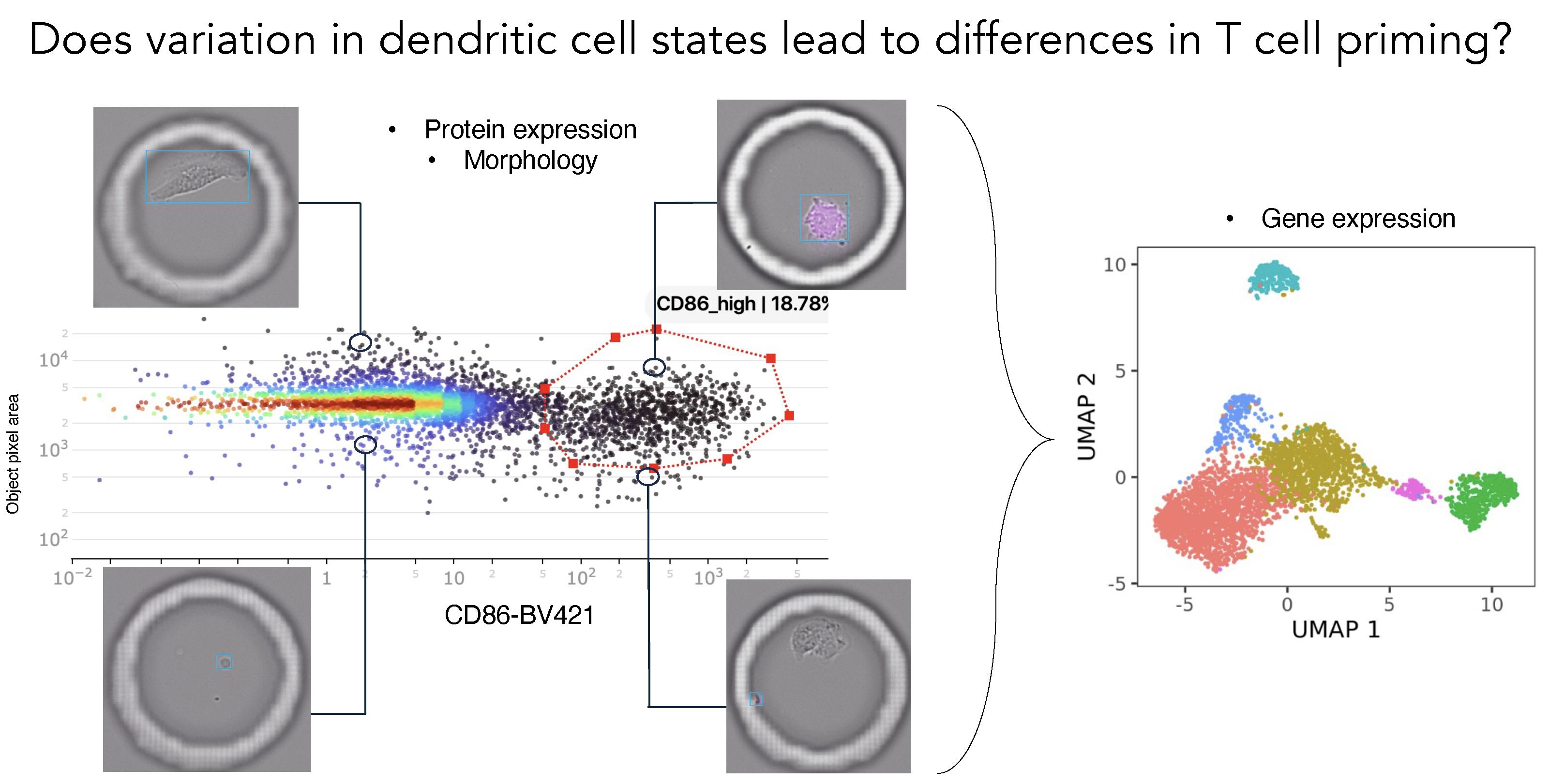
Time-lapse imaging allowed Spitzer's team to watch priming unfold over three day: as T cells progressively express activation markers and underwent clonal expansion in response to dendritic cell signals.
New Technology Opens New Questions
This work extends CRISPR screening beyond cell-intrinsic readouts (what happens inside an edited cell) to ensemble-level outcomes - how a perturbation in one cell reshapes the behavior of its interacting partners.
“These are all the building blocks that we needed to achieve to now start to do some of these really complex cell-cell interaction genetic screens with cell extrinsic readouts, which we’re really excited about”.
Because imaging, perturbation, and RNA are captured from the same interacting ensembles, the lab can follow immune responses as they unfold:
“That is now working really nicely. We can measure all of these features within individual cages and link back the phenotypes of these really impressively responding cages that capture the biology we’re after - where we see robust T cell proliferation - and link that back to the unique features of those dendritic cells that provided those activating signals.”
This makes it possible to identify which specific interactions produce robust immune responses, and to trace those outcomes back to the RNA profiles that drive them.
From Discovery to Clinical Application
With technical validation complete, Spitzer's lab is scaling to multi-gene screens with clear translational potential.
Since dendritic cell vaccines are already FDA-approved for prostate cancer, Spitzer notes: "Any genetic hits that we identify from these types of assays could be really rapidly translated into new clinical therapeutic cellular products."
The lab is also applying this approach to study T cell cytotoxicity against tumor cells, aiming to identify functional states associated with superior killing capacity and ways to genetically enhance effectiveness.
While Spitzer's focus is cancer, this framework generalizes to any systems where outcomes are defined by cell-cell interactions, including neuroscience, infectious disease, and regenerative medicine.
A Collaborative Partnership
As an early collaborator during platform development, Spitzer describes his experience with Cellanome:
"I’ve never had such a productive and close-knit collaborative interaction with a new technology company in my professional career, despite having worked on the development of a number of different technology platforms over the years."
This work, led primarily by Dr. Camilla Valente with contributions from other lab members, illustrates what becomes possible when researchers can directly measure cellular interactions that determine biological outcomes.
Explore Further
Watch the full talk to hear Matt go deeper on the experiments, data, and implications.
Hear the broader story: In our podcast conversation with Dr. Spitzer, Matt discusses his ongoing collaboration with Cellanome, computational innovations making this data analyzable, and future directions including AI-driven modeling of multicellular interactions.
Explore similar approaches in your research: Contact our team to discuss how cell-extrinsic screening could advance your work, or explore our resources on live-cell interaction studies.
