
Resources: Presentations
Multimodal Live-Cell Biology at Single-Cell Resolution: The Cellanome R3200 Platform
Presented by:
Gary Schroth, Ph.D.
Introduction
Most technologies capture only fragments of cellular behavior: a static snapshot, a transcriptomic profile, an endpoint assay. None can follow the same cells over time while simultaneously capturing what they do and what genes they express. This fundamental limitation forces researchers to infer connections between cellular phenotypes and molecular states by stitching together data from separate experiments, never seeing the complete story unfold in real time.
Dr. Gary Schroth's presentation demonstrates how the Cellanome R3200 platform addresses this challenge by unifying live-cell imaging, programmable microenvironments, and single-cell transcriptomics within a fully automated workflow. The result is what Cellanome calls cellular choreography: follow the same cells or ensembles over time, capture movement, interactions, and functional changes, then link these dynamic behaviors directly to transcriptomic profiles and sgRNA expression from the same cells.
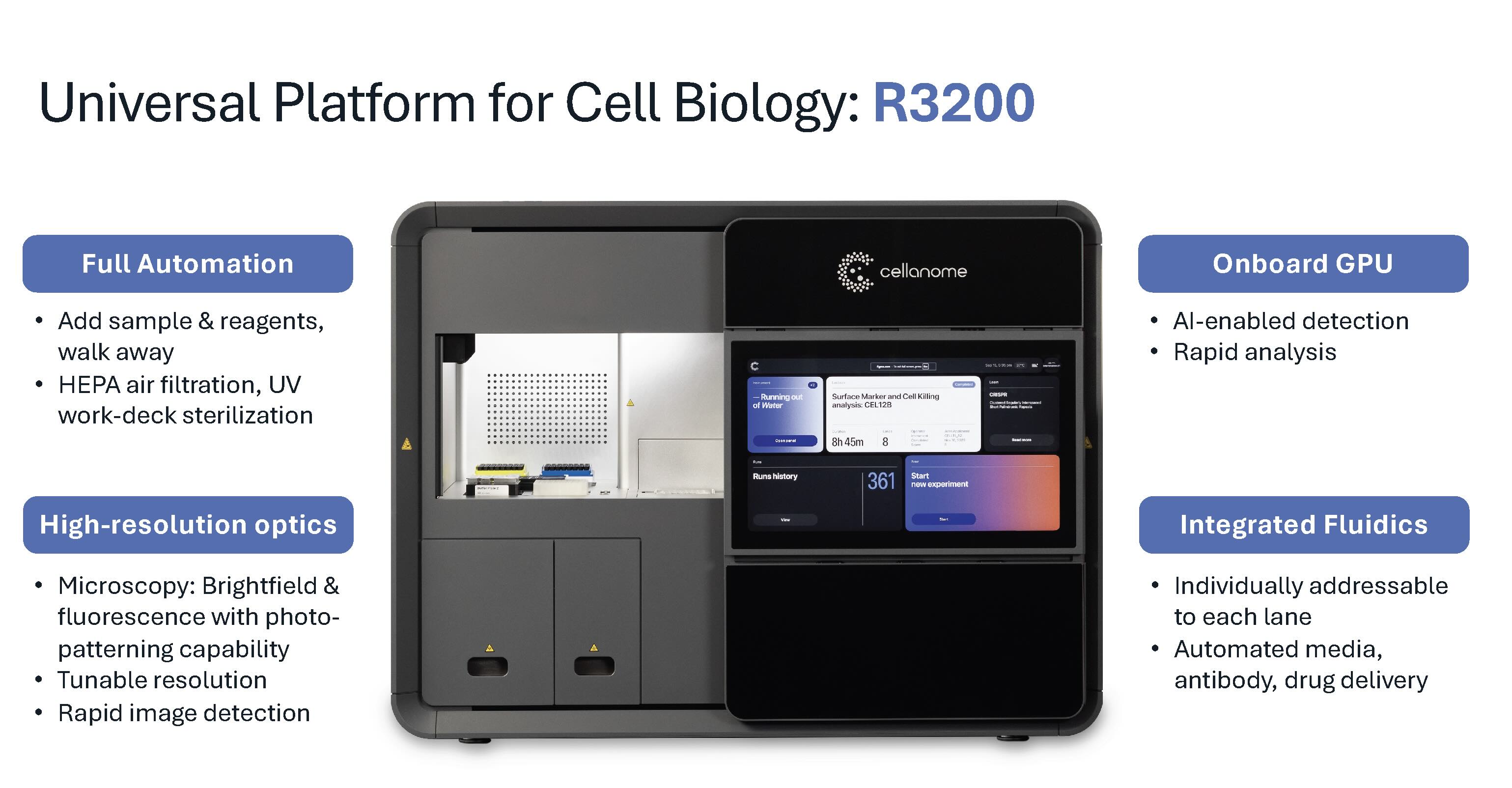
Unifying What Cells Do with What Genes They Express
The R3200 was purpose-built to integrate experimental modalities that typically require separate instruments and workflows. As Dr. Schroth explains, the platform enables researchers to "link live cell assays, we can make morphology, we can link what the cells are doing to eventually transcriptome and a CRISPR guide perturbome." This integration represents, in his assessment, "one of the only platforms, if not the only platform in the world" combining these capabilities within a single experiment.
Traditional approaches force researchers to choose: image cells over time, or profile their transcriptomes. Not both. The R3200 eliminates this trade-off. It captures live-cell dynamics through time-lapse imaging, performs functional assays during culture, and recovers single-cell transcriptomes from the same cells within programmable microenvironments. This cellular choreography reveals not just what genes are expressed, but what cells were actually doing before and during that expression.
The R3200 platform architecture seamlessly integrates sophisticated brightfield and fluorescent imaging optics, an onboard GPU for real-time image analysis, and liquid handling robotics for automated reagent delivery. These components work together to generate complete multimodal datasets without manual intervention.
“The Genius of Cellanome:” Custom Enclosures Built in Seconds
The platform's defining innovation lies in what Dr. Schroth calls "the genius of Cellanome": a patented photopatterning system that creates custom, biocompatible CellCage™ enclosures around selected cells in real time. The process begins when cells flow into the flow cell along with a proprietary monomer chemistry. Within 2-3 seconds per image, the system captures an image, performs AI-enabled analysis to identify cells of interest, and uses a digital micromirror device (DMD) to project patterned light around selected cells. This light triggers polymerization, forming semi-permeable walls that span from floor to ceiling of the flow cell and create individualized compartments. The entire enclosure process for a full lane takes 10-15 minutes.
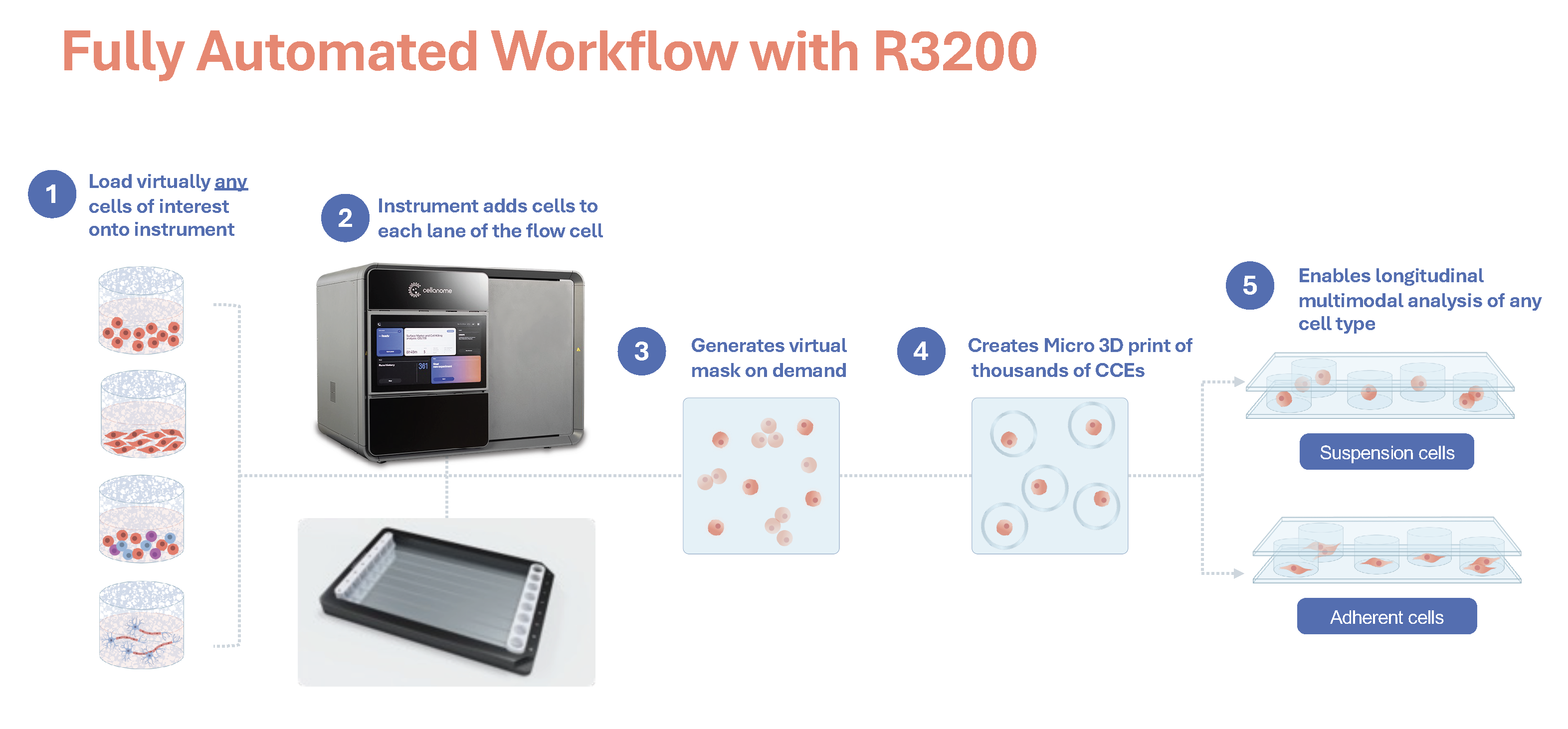
"Whatever you can imagine, we can print on the flow cell," Dr. Schroth notes. The programmability extends beyond simple circular enclosures to custom geometries tailored to experimental needs, such as window cages for studying migration and maze-like structures for directional guidance. The polymer walls maintain spatial separation while remaining permeable to biochemicals. This allows reagents, including large molecules like antibodies, media components, and signaling factors, to freely access cells while preventing mixing between enclosures.
Critically, the platform accommodates adherent cells in their native growth state, a capability Dr. Schroth emphasizes as key differentiation. "Most cells are adherent. They prefer to be attached to a surface, and we allow that to happen in our platform." Most single-cell platforms require cells to be maintained in suspension, which means they are dissociated from their natural attachment points and prevented from exhibiting their native morphology and behavior.This compromise fundamentally alters cellular biology for the vast majority of cell types, including neurons, epithelial cells, fibroblasts, and adherent cancer lines. The R3200 takes a different approach: flow cells can be pre-coated with fibronectin, PLO-laminin, or other extracellular matrix proteins, enabling cells to attach, spread, and exhibit the morphology and behavior they would display in native tissue or standard adherent cell culture.
After live-cell imaging and functional assays, cells are lysed in place. RNA binds to barcoded capture probes on the flow cell surface, undergoes conversion to cDNA, and is released for sequencing. Barcodes enable computational assignment of every transcript back to its originating enclosure, linking each cell’s molecular profile to its observed behavior. This provides the complete cellular choreography from start to finish.
From Hours to Weeks: Capturing Biology at Every Timescale
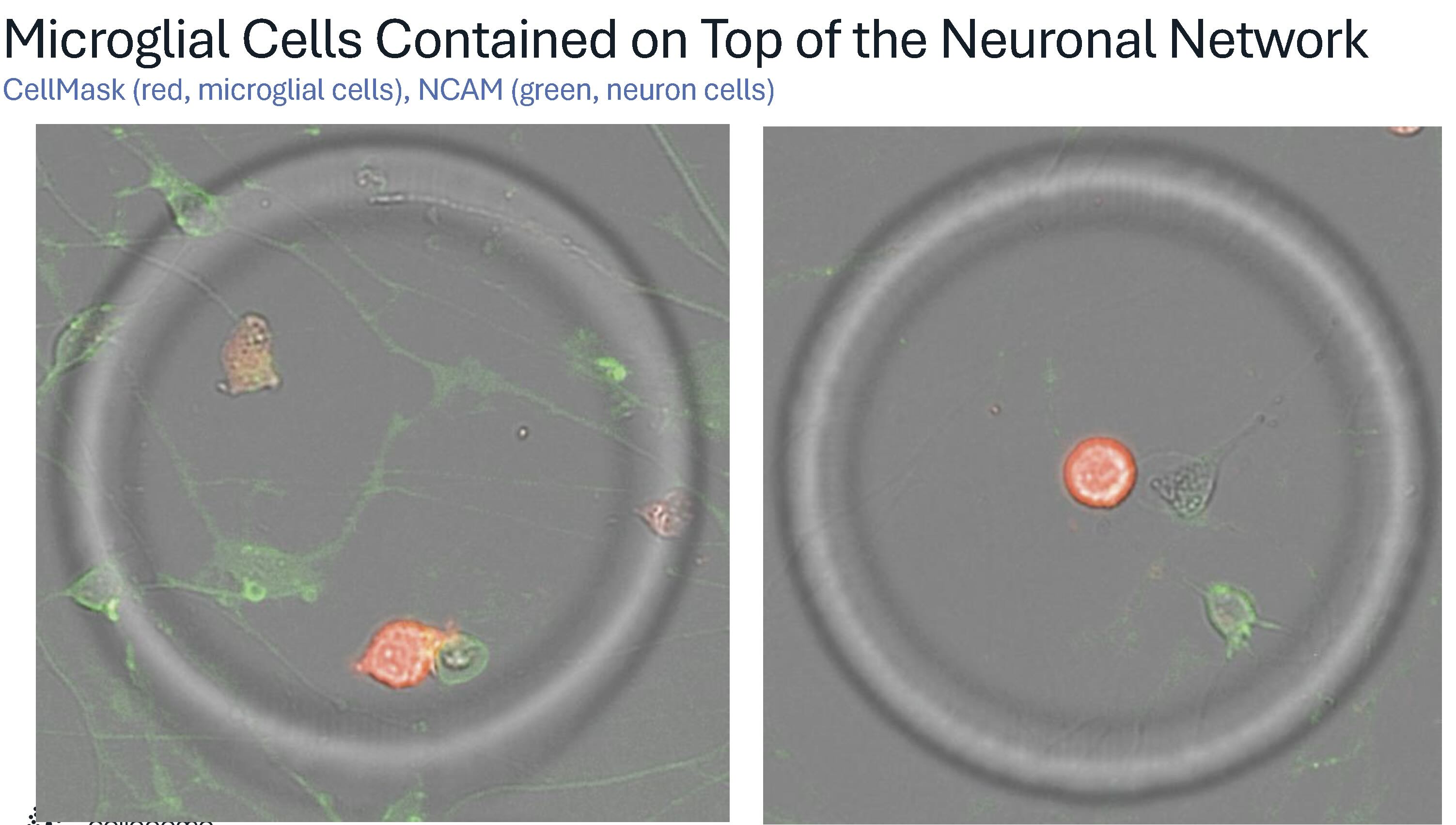
These technical capabilities translate into diverse experimental applications across multiple biological systems and timescales. One recent demonstration captures this scale and flexibility in action. Just weeks before the presentation, primary neurons were cultured on PLO-laminin-coated flow cells for two weeks, forming dense, highly branched networks that "look almost three-dimensional." Microglial cells were then introduced onto this neuronal bed and confined within CellCage enclosures, creating individualized microenvironments. Dr. Schroth describes these as “a little playpen for them on top of this bed of neurons.”
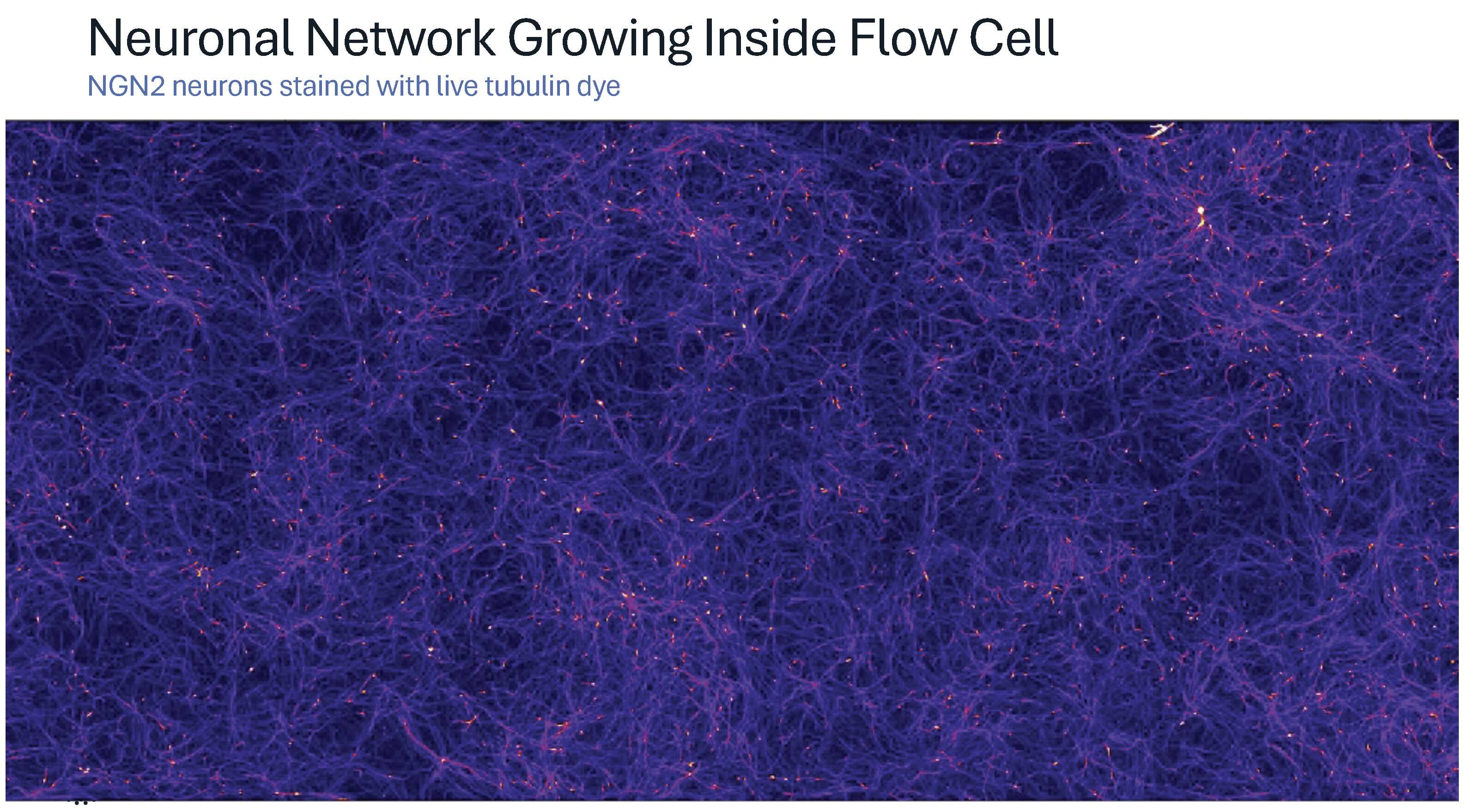
Time-lapse imaging captured the full choreography of microglial behaviors: active patrolling, phagocytosis of debris, cell division, and interactions with neuronal processes. In one remarkable sequence, a microglial cell consumed so much debris "that it divides into two cells." Another sequence revealed what Dr. Schroth suspects is neuronal pruning in action: "This glial cell was reaching down there and sort of seems to snip the neuron. And you can see it recoil after it gets snipped."
The scale of parallel observation distinguishes this approach from conventional imaging. "This is one of thousands and thousands of cages," Dr. Schroth notes. "Each cage has a story like this going on inside of it." A single lane generates more than 5,000 individual enclosures. Each functions as an independent microenvironment with its own time-lapse image of cellular dynamics, followed by transcriptomic profiling, capturing the full performance of cellular choreography at an unprecedented scale.
Other demonstrations span diverse biological systems and timescales. T-cell-mediated killing assays capture cytotoxic dynamics over hours, with time-lapse imaging revealing how individual T cells attack target cells and trigger death signals. Induced pluripotent stem cell differentiation tracked over weeks demonstrates the platform's ability to monitor gradual cell fate transitions while profiling transcriptomic changes that drive differentiation. Custom cage geometries enable migration studies through window cages and perturbation screens with functional readouts. These examples illustrate the platform's applicability across immunology, neuroscience, stem cell biology, and oncology research.
An Open, Customizable System for Discovery
Dr. Schroth frames the R3200 as a "universal toolkit for cell biology," emphasizing the platform's experimental flexibility. The combination of programmable enclosure geometries, automated multi-day adherent cell culture with reagent delivery, time-lapse imaging across multiple fluorescent channels, and downstream transcriptomic analysis enables custom assay design tailored to specific biological questions.
This openness has fostered rapid adoption and innovation. "In the course of the last year or so we've had 15 different posters at 15 international conferences together with collaborators," Dr. Schroth reports. "It's amazing what people come up with to do with a technology that's so open and kind of customizable as ours is." The expanding portfolio demonstrates how capturing the full cellular choreography, linking dynamic behaviors to molecular programs, opens experimental possibilities previously inaccessible with conventional technologies. From studying rare cell state transitions to mapping the choreography of how cellular phenotypes emerge from genetic perturbations, researchers can now ask questions about the temporal coordination of cellular processes that were impossible to address before.
Conclusion
The Cellanome R3200 platform addresses a critical need in cell biology: connecting dynamic cellular behaviors to the molecular programs that drive them. By integrating long-term live-cell imaging, programmable microenvironments, functional assays, and single-cell transcriptomics within an automated workflow, the R3200 enables researchers to capture cellular choreography, which is the full performance of how cells move, interact, change, and express genes over time. From immune cell dynamics captured over hours to neuronal network formation tracked over weeks, the platform's flexibility supports diverse experimental timescales and biological systems, revealing the temporal patterns and coordinated responses that define living cells.
Explore how the R3200 can advance your research. View platform applications or contact our team to discuss how multimodal, longitudinal single-cell analysis can address your specific research questions.
