
Resources: Presentations
Multimodal and Context-Aware Profiling for Disease Modeling: Studying Cells in Context
Presented by:
Faranak Fattahi, Ph.D.
Introduction
Drug discovery traditionally follows a gene-centric approach: identify a disease-causing mutation, determine which protein is dysfunctional, and design a therapeutic to fix it. This works well for monogenic diseases or cancers driven by a single oncogene. "But a lot of times in neurological diseases, that process is so much more complicated," explains Dr. Faranak Fattahi, Associate Professor of Cellular and Molecular Pharmacology at the University of California, San Francisco. There isn't one mutation driving disease in all patients. "What we are looking for is more holistic phenotyping of the cells under the disease state to figure out what goes wrong at the cellular behavior and state level and then come up with a therapeutic strategy to fix that overall phenotype," she explains.
Dr. Fattahi's research demonstrates how Cellanome's R3200 platform addresses this challenge through two complementary approaches: multimodal phenotyping and context-aware profiling. These capabilities enable researchers to measure multiple cellular behaviors simultaneously within physiologically relevant cellular contexts.
The Challenge: Integrating Fragmented Data
"Historically, you have to take one strategy or one technology at a time," Dr. Fattahi explains. Gene expression profiling in one population, proliferation or migration in another, electrical activity in a third, synaptic activity and interactions in a fourth. "In the past couple of decades, you have to measure one of these things at a time and try to understand one step at a time, what could be going wrong and try to integrate all of that information into a holistic understanding of what happens in the cells."
This sequential approach creates inherent limitations. Researchers must culture separate cell populations for each assay, then attempt to integrate datasets generated under different conditions and timescales. The result is a fragmented picture of disease mechanisms, where dynamic relationships between cellular phenotypes remain obscured. "But with Cellanome," she says, "you could look at all of that together in one step."
The platform enables integrated workflows where neuronal populations, including 3D neurospheres containing thousands of neurons, undergo longitudinal live-cell imaging while being cultured in CellCage™ enclosures. Researchers track morphology, synaptic markers, and calcium dynamics over time, then profile the same cells transcriptomically at experimental endpoints. This approach directly correlates dynamic imaging data with gene expression states.
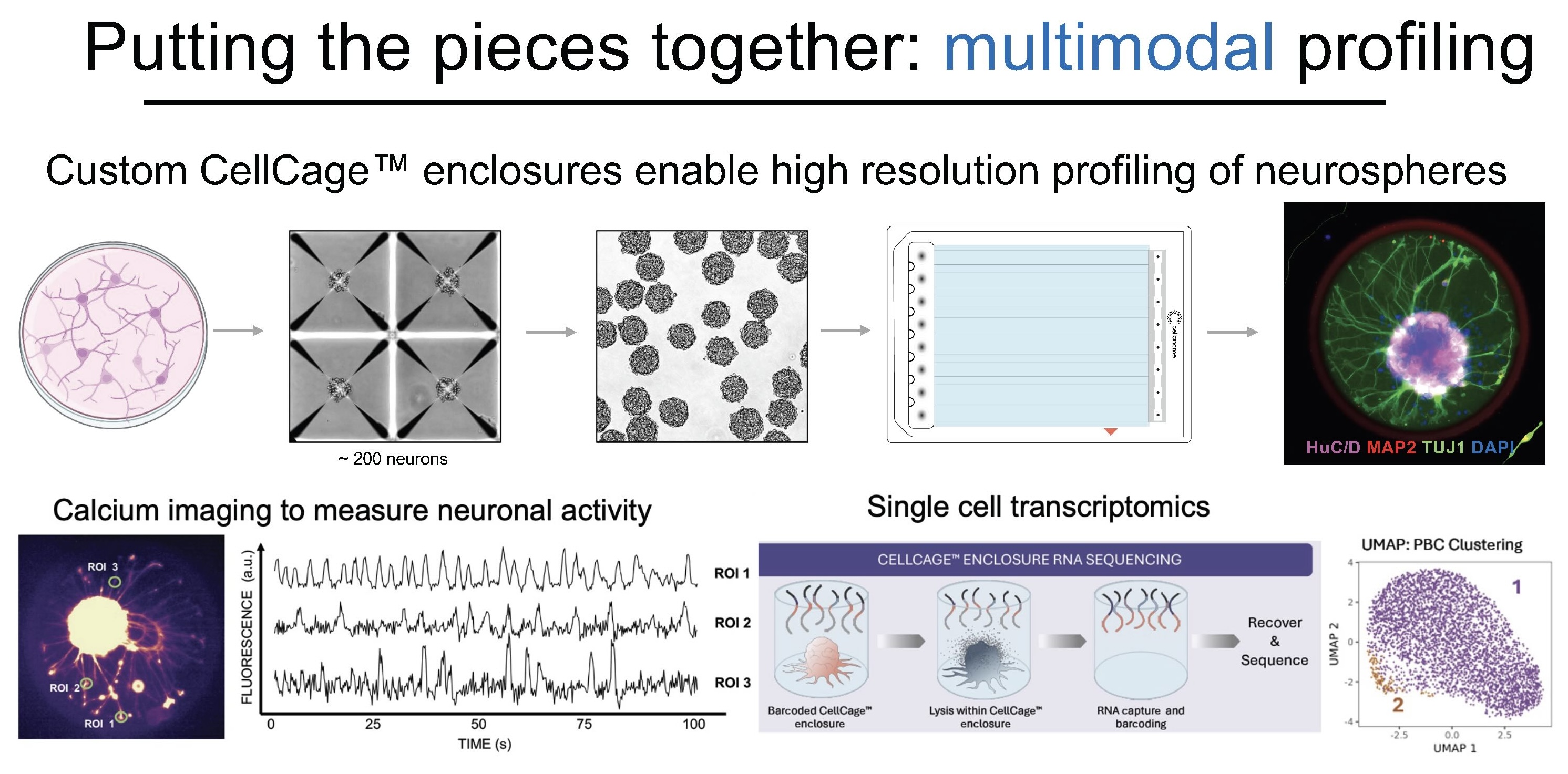
Multimodal Phenotyping in Practice
Dr. Nick Elder, formerly in the Fattahi lab and now at Cellanome, demonstrated this integrated approach by differentiating three types of neurons from stem cells using different differentiation protocols. Neuronal cultures grown as neurospheres were loaded into custom CellCage™ enclosures with windowed designs that allow axonal processes to extend beyond the cage boundaries. Elder profiled all three neuronal populations on the R3200 platform, correlating imaging data, specifically blue fluorescent protein expression, directly with gene expression patterns.
"This allows you to correlate very complex measurements of transcription to behaviors that are very dynamic and long-term," Dr. Fattahi notes. The platform captures not just endpoint snapshots but the temporal evolution of cellular states, linking transcriptional programs to observable phenotypic changes.
Why Context Matters: The Extended Phenotype
Multimodal profiling captures complexity within cells; context-aware profiling captures complexity between them. "You can have cells in their normal physiological interaction with other surrounding cells in their neighborhood and look at how disruptions in one process in one cell type leads to changes in the state in another cell type," Dr. Fattahi explains.
This matters because "a lot of these neurological diseases that we're interested in studying are not just disruptions of one cell function. They're disrupted in the tissue." A glial problem leads to disruptions in neurons. Immune cells disrupt the vasculature. The vasculature causes problems in neuronal signaling. "We want to be able to measure all of these things in that true biological context."
Context-aware profiling enables what Dr. Fattahi calls the "extended phenotype," measuring how perturbations in one cell type affect neighboring cells they interact with. "If you co-cage them together, you have yellow cells and pink cells together. You can perturb the yellow cells, but then that change in yellow cells is going to change the pink cells too, and you can measure those phenotypes as well."
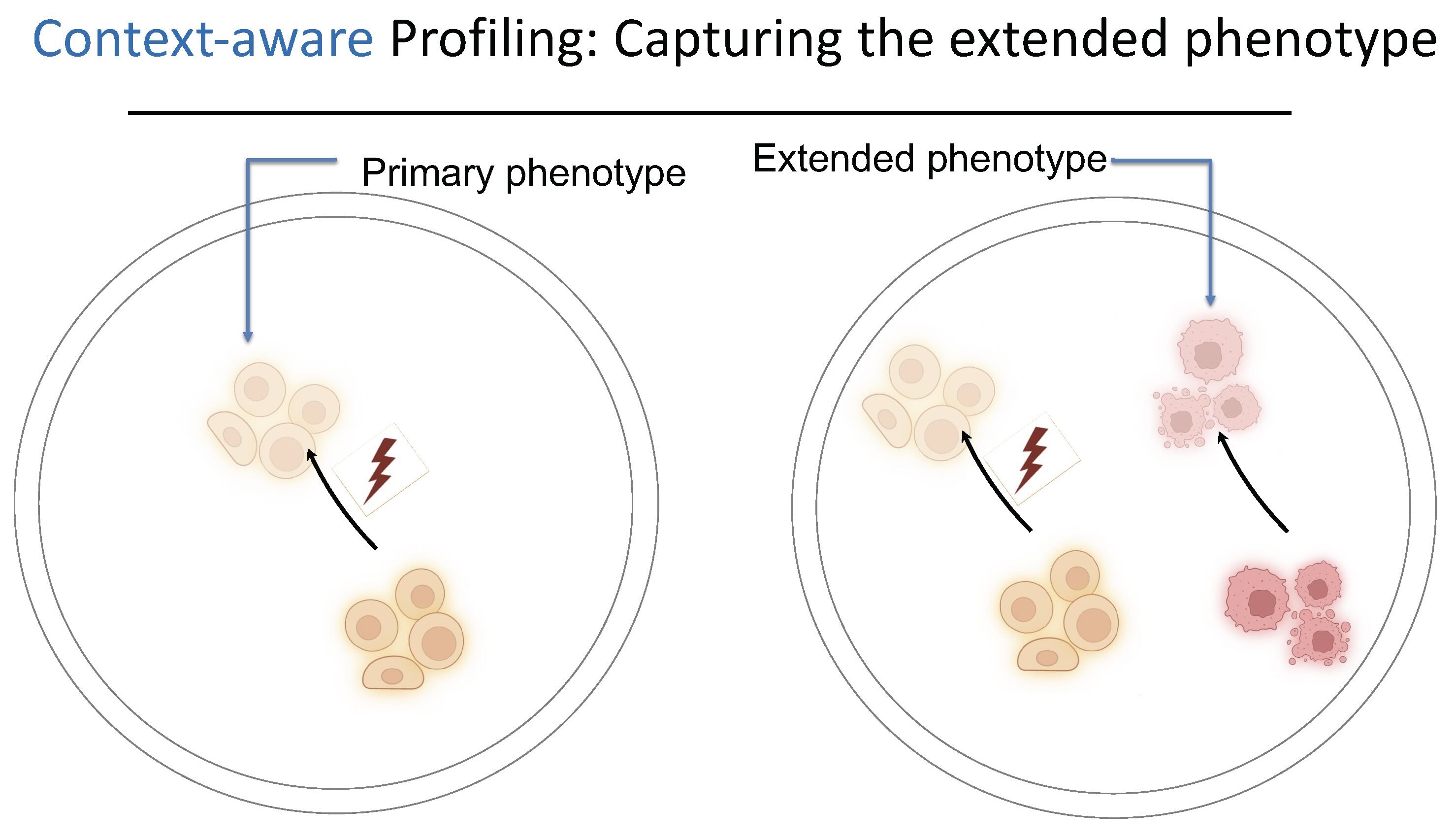
This capability addresses a fundamental limitation of current pooled CRISPR screening approaches. "This is something that's unlocked with the Cellanome platform and is currently impossible in pooled CRISPR screens," Dr. Fattahi notes. "With the traditional methods, you're measuring the outcome of your perturbation in the cell that is perturbed, but you're not able to measure that in other cells that it's interacting with."
Measuring How Cells Influence Each Other
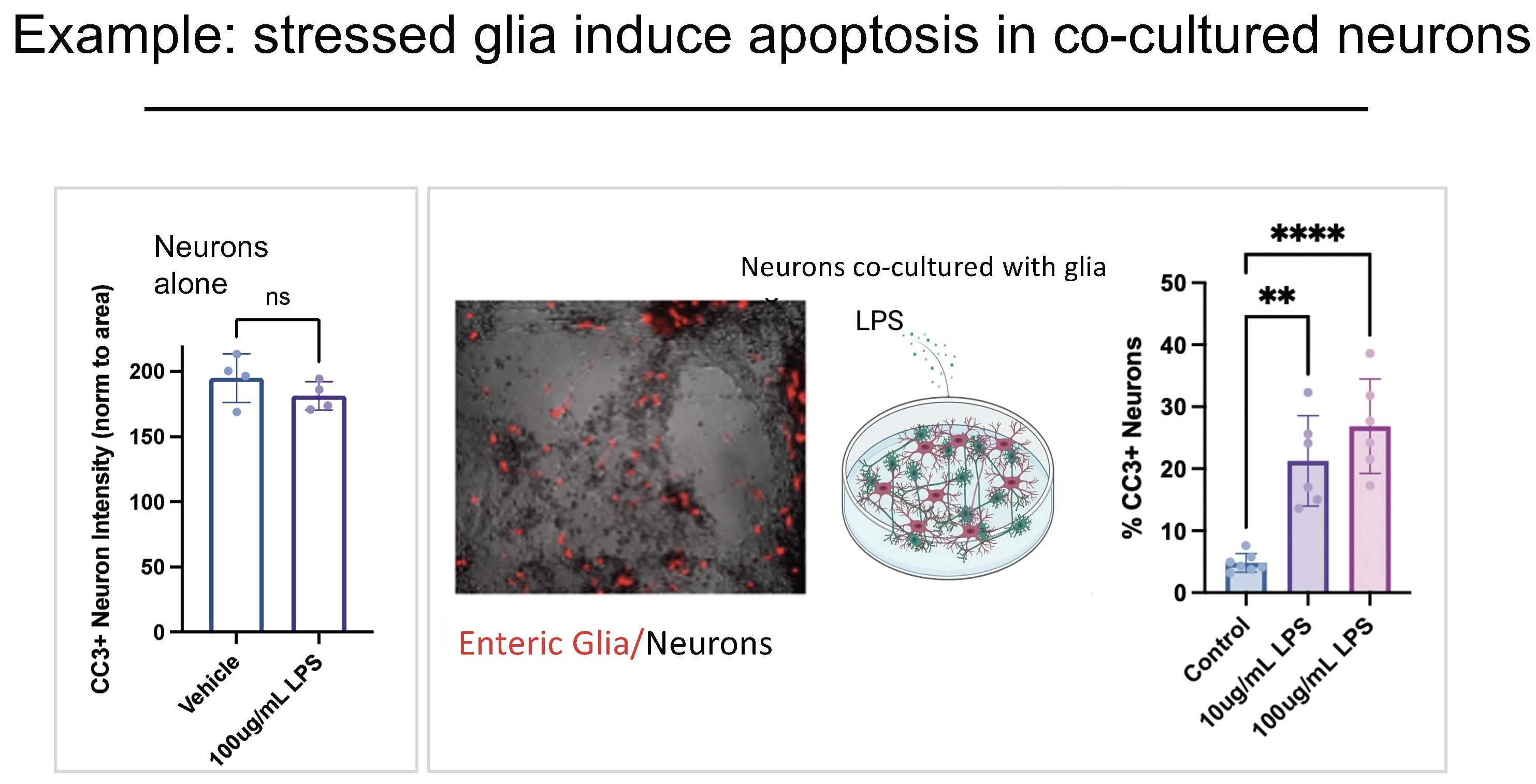
Dr. Fattahi's team demonstrated context-aware profiling through two experimental systems that reveal how cellular stress propagates through cell-cell interactions.
In the first example, the team co-cultured enteric neurons (neurons in the gut) with enteric glial cells. When neurons are exposed to LPS (an inflammatory stressor) alone, they survive. But when co-cultured with glia and then exposed to LPS, the neurons die. "This suggests that there's something coming from these glial cells under stress that causes neurotoxicity and induces damage in the neurons," she explains.
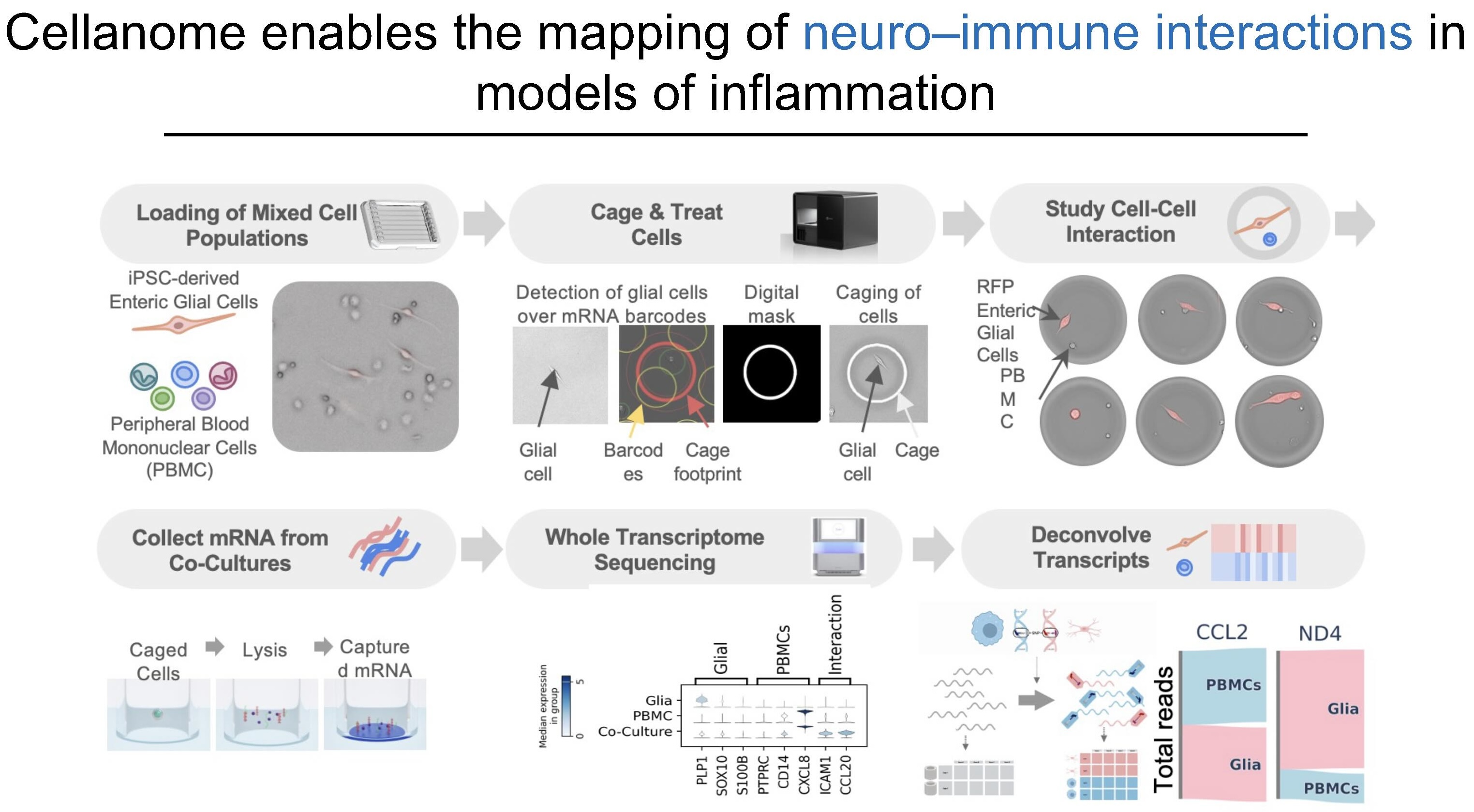
The second example explores neuro-immune interactions. Dr. Fattahi's team co-cultured enteric glial cells with PBMCs (peripheral blood mononuclear cells, immune cells from blood) from different donors. Using natural genetic variants to deconvolve which RNA came from which cell type, they found certain markers aren't expressed when either cell is cultured alone. However, "when they are co-cultured together, now all of a sudden they express these markers that suggest that there's some sort of communication between the two cell types."
These experiments reveal phenotypes that only emerge in co-culture, capturing disease-relevant mechanisms that would remain invisible in traditional single-cell-type assays.
From Data to Predictive Models
Dr. Fattahi envisions using multimodal phenotypes paired with transcriptional profiles, all measured in defined cellular contexts, to train models that predict how perturbations propagate through cell-cell interactions.
The approach starts with iPSC-derived neurons, glia, and immune cells. After inducing disease states, researchers run large-scale perturbation screens with genetic modifications or biologics. For each perturbation, they measure dynamic morphology, secretion, and gene expression from interacting cells, then train models using this data to predict how therapeutic interventions in one cell type affect the cells they interact with.
The goal extends beyond single-cell therapeutic targets. If stressed glia kill neurons, can researchers predict which interventions in glia rescue neuronal survival? If immune cells activate glia, which modulators prevent that cascade? For diseases emerging from multi-cell interactions, models trained on multimodal, context-aware data reveal therapeutic strategies that single-cell approaches miss.
A Path Forward for Complex Disease
Dr. Fattahi's research demonstrates how integrated multimodal and context-aware profiling addresses the complexity of neurological disease by measuring cellular behaviors in their native biological contexts. This approach enables next-generation predictive models for therapeutic discovery, moving from single-cell targets to understanding how interventions propagate through multi-cellular disease mechanisms.
Explore how the R3200 platform can advance your disease modeling research. Contact us to discuss applications in neuroscience, immunology, and complex disease systems, or view our application resources to see additional examples of multimodal single-cell profiling.
